|
MitoPedia: SUIT |
MitoPedia O2k and high-resolution respirometry:
O2k-Open Support
- MitoPedia: SUIT - Substrate-uncoupler-inhibitor titration protocols
- » The SUITbrowser helps to find the best SUIT protocol for your research questions.
- » Introduction to SUIT protocols
Introduction to SUIT protocols
SUIT: Substrate-uncoupler-inhibitor-titration
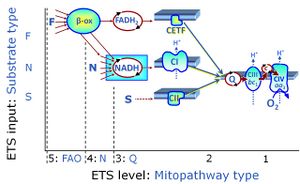
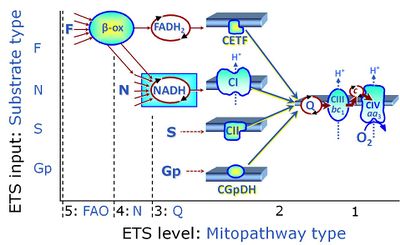
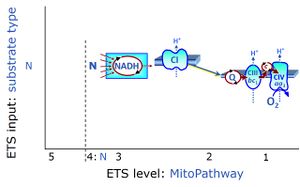
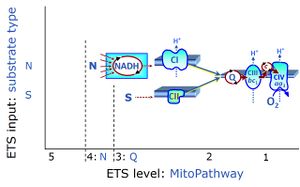
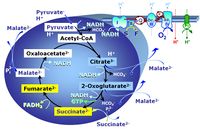
- SUIT protocols are applied to mitochondrial preparations. Categories of SUIT protocols include all fuel substrate types involved and list in parentheses the N-type substrates, independent of the sequence of titrations, e.g. NS(GM), NS(PM), FNSGp(PGM). The coupling/patwhay control diagram in the Figure shows a protocol of SUIT category FNS(PM).
- Coupling-control protocols (CCP) are a special case of SUIT protocols primarily applied to living cells (ce; ceCCP), typically with a sequence of ROUTINE respiration (R), optionally oligomycin-induced LEAK respiration (L), uncoupler-stimulated ET capacity (E), and inhibitor-induced residual oxygen consumption (Rox). Cell-membrane permeable substrates (glucose, pyruvate) may be applied to evaluate their effect on cell physiology, and cell-membrane impermeable substrates (succinate, ADP) are titrated as a cell viability test to assess the fraction of dead cells (dce) with permeable cell membranes and functional mitochondria. It is important to provide information on the composition of the respiration medium, particularly distinguishing media with exogenous substrates and high Ca2+ concentration (culture media) and mt-respiration media with Ca2+ chelators (e.g. MiR05) excluding respiratory substrates, when respiration is strictly based on endogenous ET-pathway substrates.
- Educational: The library of SUIT protocols is developed to introduce the basic concept by simple coupling/pathway control diagrams.
- Practical: The SUIT protocols must meet the detailed practical requirements to provide a complete guide through the titration experiment, defining every titration step (see DL-Protocols).
- Compatible: The systematic name of a SUIT protocol must be unique for each variation within a category of SUIT protocols, compatible with a database.
- Fast: The terminology should help to recognize the essentially features of a SUIT protocol sufficiently fast, but detailed explanations are provided in the definitions, explanations and discussions.
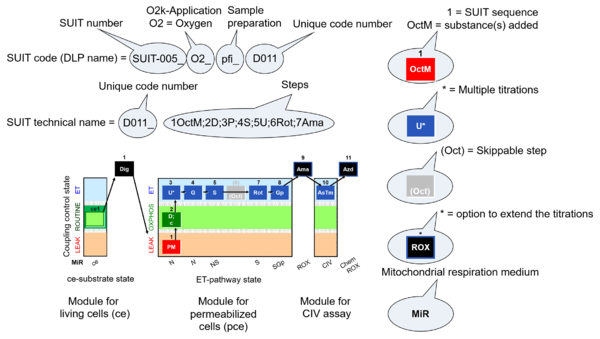
- SUIT sequence - sequence of titration steps: Each titration step on coupling control and pathway control starts with a sequential number, followed by the acronym of the substance titrated. Some respiratory states specifically require multiple titrations of the same substance for evaluation of the optimum concentration. Such multiple titrations are indicated by an asterisc following the acronym of the substance.
- 5U* - stepwise titration of an uncoupler to measure ET capacity as the noncoupled state of maximum flux at optimum uncoupler concentration.
- Quality control: Titration steps related to quality control (e.g. cytochrome c test) may or may not be included in a selected state of a SUIT protocol, are not considered in the categories of SUIT protocols, and do not have a new number in the name of the SUIT protocol but maintain the number of the particular coupling/pathway control state.
- Flexibility in DL-Protocols: Fixed sequence of events and marks can be changed (Skip/Added) in a SUIT protocol by the user (to be provided in the next DatLab version). Editions of concentrations and titration volumes of injections in a specific DL-Protocol are already available as user-specific DL-Protocol [File]\Export\DL-Protocol User (*.DLPU).
- indicates in SUIT protocols the option to extend the titrations by (1) inhibition of electron transfer (typically by antimycin A, Ama) to induce ROX, optionally followed by an assay of cytochrome c oxidase activity, with sequential titration of ascorbate and TMPD (AsTm) and azide (Azd) for evaluation of the chemical O2 background due to autoxidation reactions.
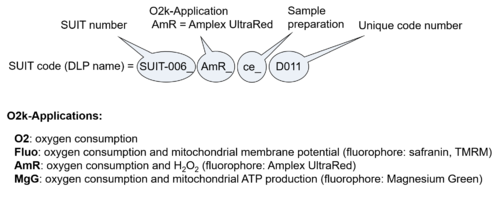
- O2k-Application: Displays the O2k-Applications of a specific SUIT protocol.
- Modular approach: Select your DLP protocol from our archive of recommended SUIT-A library. The modular approach is designed to make easier to resolve your scientific question using protocols that guide you step by step.
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Library of SUIT protocols
SUIT A Respirometry: recommended protocols
- »A: MitoPedia: SUIT A - SUIT protocols used for cohort studies, available as DL-Protocols with DatLab 7. SUIT A protocols have a unique D### code: e.g. SUIT-008 O2 pfi D014).
| SUIT number | Short name | Description/Reference | Coupling/pathway control diagram |
|---|---|---|---|
| SUIT-001 | RP1 | Doerrier 2018 Methods Mol Biol | 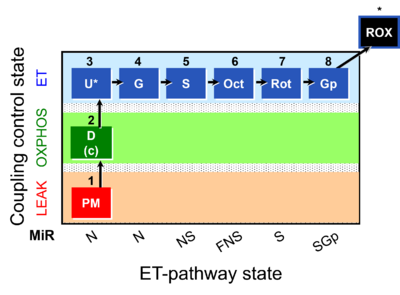 |
| SUIT-002 | RP2 | A: Doerrier 2018 Methods Mol Biol | 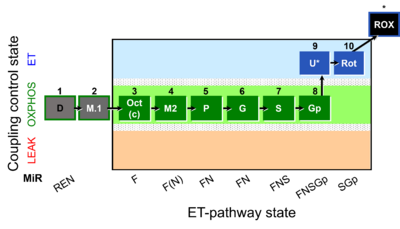 |
| SUIT-003 | CCP-ce | A: Coupling-control protocol, living cells »MiPNet08.09 CellRespiration Bioblast pdf | 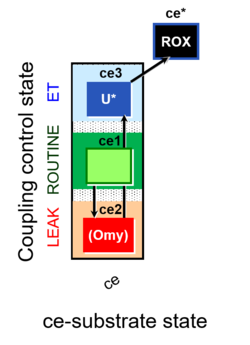 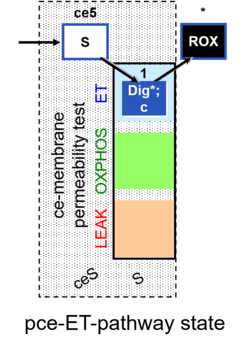 |
| SUIT-004 | RP1-short | 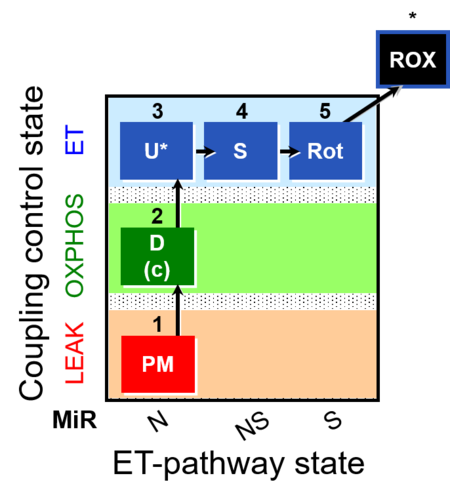 | |
| SUIT-005 | RP2-short | A: when malate-anaplerotic activity is zero | 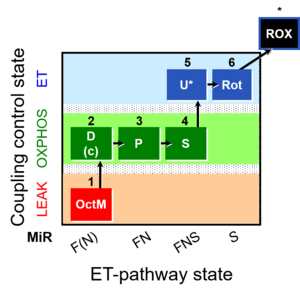 |
| SUIT-006 | CCP-mtprep | A: Coupling-control protocol, mtprep | 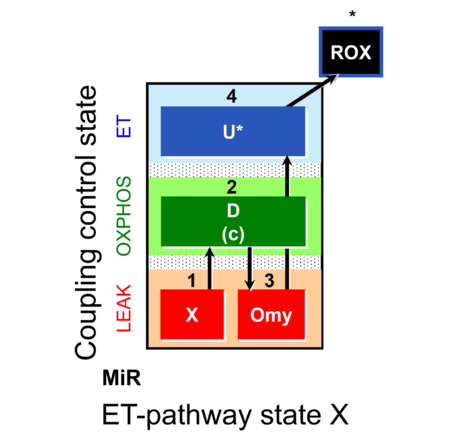 |
| SUIT-007 | Glutamate anaplerosis | A: Glutamate anaplerotic pathway | 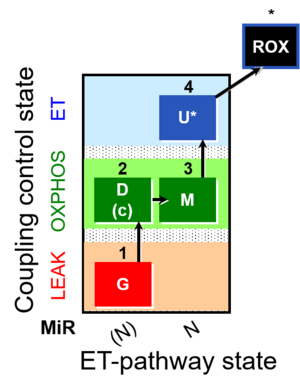 |
| SUIT-008 | PM+G+S_OXPHOS+Rot_ET | A: Additivity between the N- and S-pathway in the Q-junction | 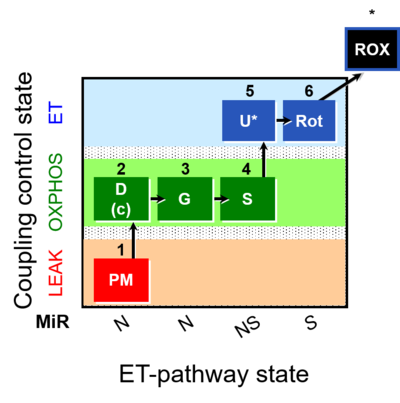 |
| SUIT-010 | Digitonin test | A: Optimization of digitonin concentration for pce | 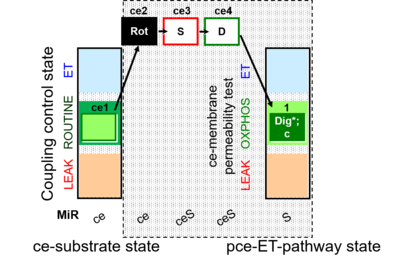 |
| SUIT-011 | GM+S_OXPHOS+Rot_ET | A: Maximum mitochondrial respiratory capacity (OXPHOS with NS substrates) and coupling/pathway control | 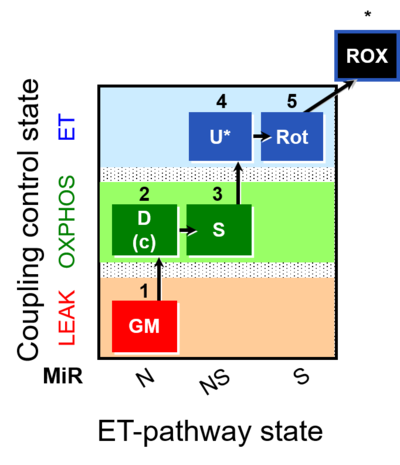 |
| SUIT-012 | PM+G_OXPHOS | A: Coupling control (L- P- E) with NADH-linked substrates (PM and PGM) | 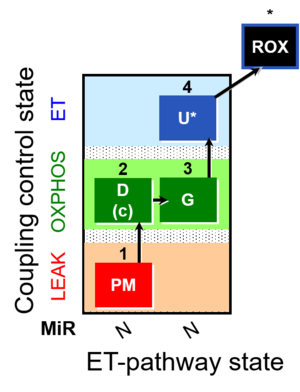 |
| SUIT-014 | GM+P+S_OXPHOS+Rot_ET | A: Cells or tissue types that display a preference for GM over PM to support NADH-linked respiration | 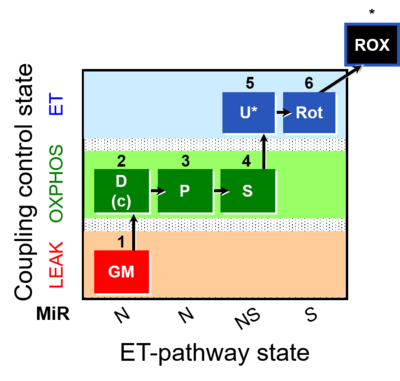 |
| SUIT-015 | F+G+P+S_OXPHOS+Rot_ET | A: F-pathway in LEAK state and OXPHOS state | 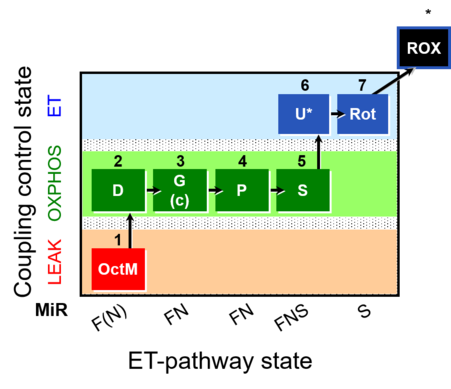 |
| SUIT-016 | F+G+S+Rot_OXPHOS+Omy | A: F-pathway in LEAK state and OXPHOS state | 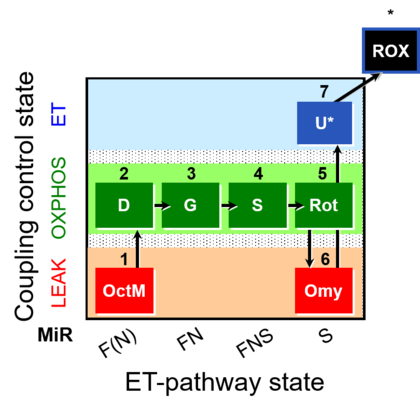 |
| SUIT-017 | F+G+S_OXPHOS+Rot_ET | A: | 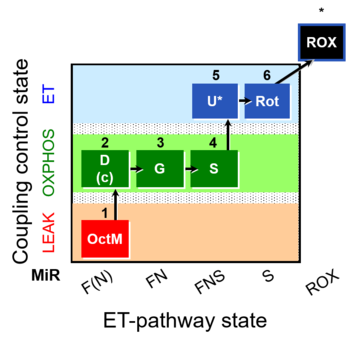 |
| SUIT-019 | Pal+Oct+P+G_OXPHOS+S+Rot_ET | A: | 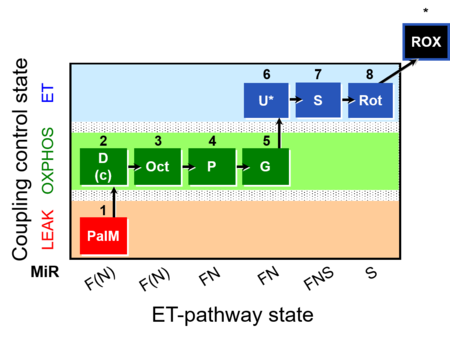 |
| SUIT-020 | PM+G+S+Rot_OXPHOS+Omy | A: simultaneous determination of O2 flux and mt-membrane potential | 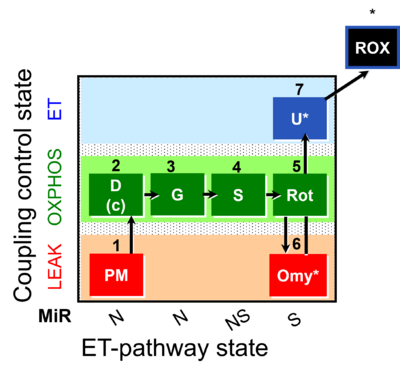 |
| SUIT-021 | OXPHOS (GM+S+Rot+Omy) | A: simultaneous determination of O2 flux and mt-membrane potential | 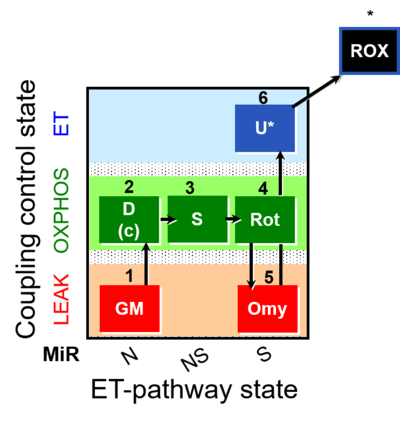 |
| SUIT-022 | AOX (ce CN+SHAM) | A: Determination of the respiration due to the alternative oxidase pathway | 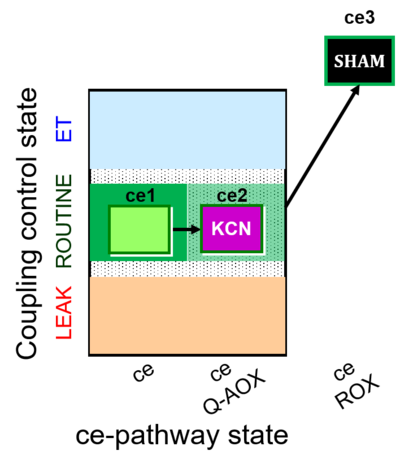 |
| SUIT-023 | AOX-ce SHAM+CN | A: Determination of respiration through the CIII-CIV pathway | 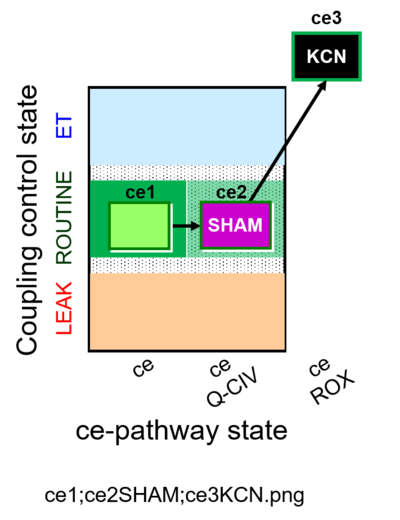 |
| SUIT-024 | ATPase (PM) | A: Determination of ATPase activity in mitochondrial preparations. | 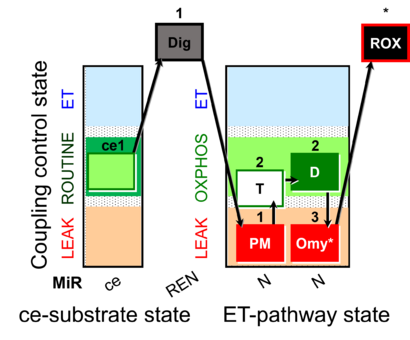 |
| SUIT-025 | OXPHOS (F+M+P+G+S+Rot) | 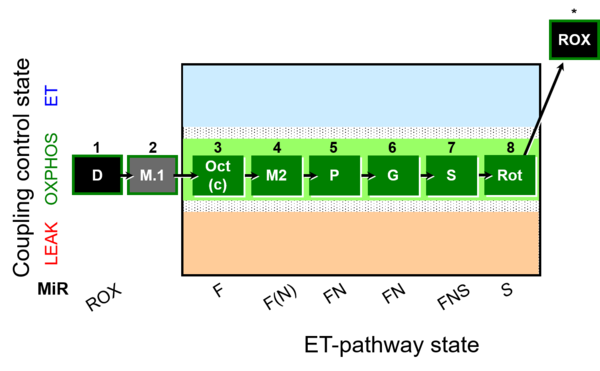 | |
| SUIT-027 | Malate anaplerosis | A: Malate anaplerotic pathway | 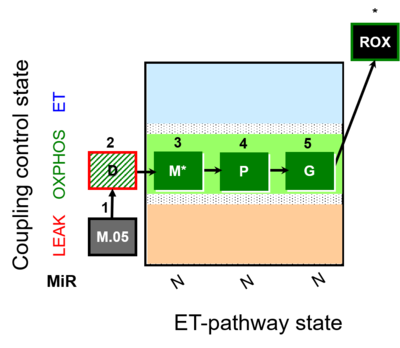 |
SUIT A FluoRespirometry: recommended protocols
- »A: MitoPedia: SUIT A - these are SUIT protocols used for cohort studies, and are available as DL-Protocols with DatLab 7. SUIT A protocols have a unique D## code: e.g. SUIT-008 O2 pfi D14).
| SUIT number | Description/Reference | Coupling/pathway control diagram |
|---|---|---|
| SUIT-006 | A: Coupling-control protocol, mtprep |  |
| SUIT-013 | A | 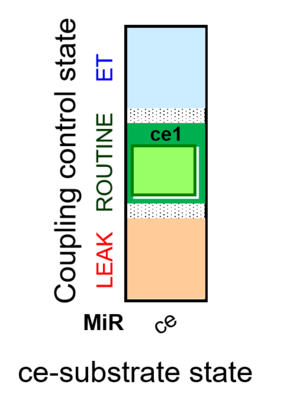 |
| SUIT-018 | A: A protocol for oxygen dependence of O2 flux and H2O2 flux in isolated mitochondria,tissue homogenate or permeabilized cells | 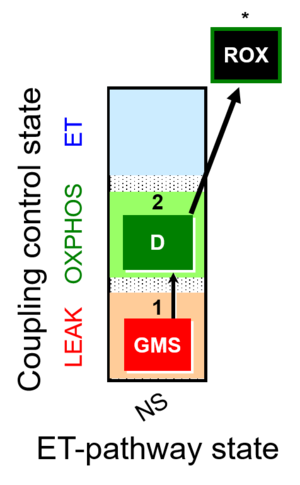 |
| SUIT-020 | A: simultaneous determination of O2 flux and mt-membrane potential |  |
| SUIT-021 | A: simultaneous determination of O2 flux and mt-membrane potential |  |
| SUIT-026 | A Short protocol to study RET-related H2O2 production | 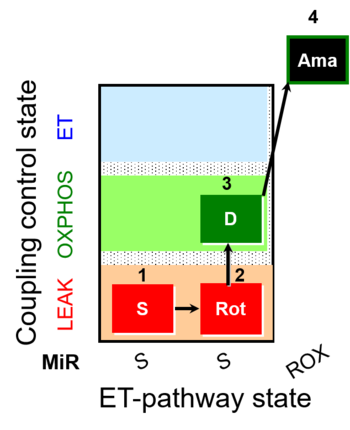 |
SUIT A: all recommended protocols
- » A: MitoPedia: SUIT A - SUIT protocols used for cohort studies
SUIT B: published protocols
- » B: MitoPedia: SUIT B - these are published SUIT protocols, not considered as level A.
SUIT C: exploratory protocols
- » C: MitoPedia: SUIT C - these are SUIT protocols used for exploratory research.
All SUIT protocols
- » Library of SUIT protocols: all SUIT protocols
| SUIT code | O2k-Application | Coupling/pathway control diagram | SUIT number | Description/Reference |
|---|---|---|---|---|
| 1OctM;2D;3PG;4S;5U;6Rot- | O2 | 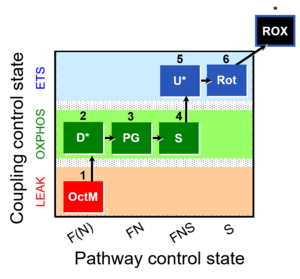 | C MiPNet18.13 IOC84 Alaska | |
| 1PGM;2D;3S;4Rot;5U- | O2 | 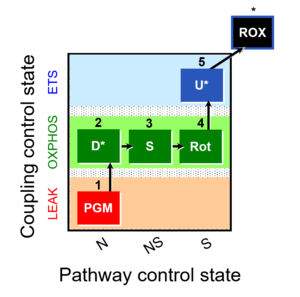 | C | |
| 1PGM;2D;3U;4S;5Rot- | O2 | 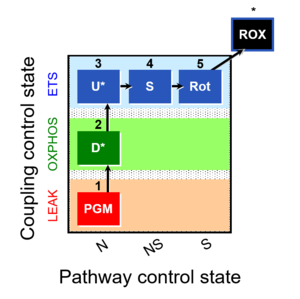 | C MiPNet18.13 IOC84 Alaska | |
| 1PM;2D;3G;4U;5S;6Rot- | O2 | 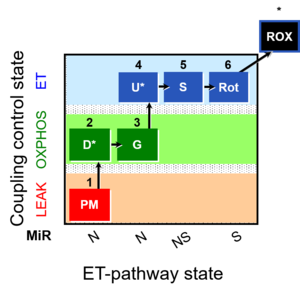 | B Lemieux 2011 Int J Biochem Cell Biol | |
| SUIT-001 |  | Doerrier 2018 Methods Mol Biol | ||
| SUIT-001 O2 ce-pce D003 | O2 | 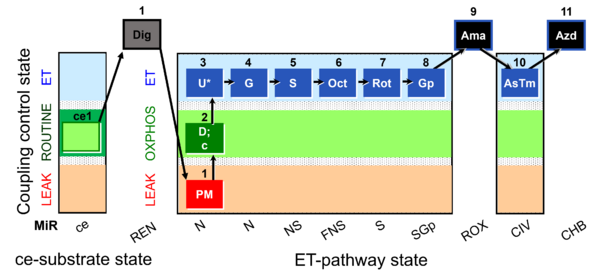 | D003_ce1;1Dig;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd | A - SUIT-001 - SUIT reference protocol Reference protocol RP1 for living cells (ce, ROUTINE respiration) and cells permeabilized in the respirometric chamber (pce) |
| SUIT-001 O2 ce-pce D004 | O2 | 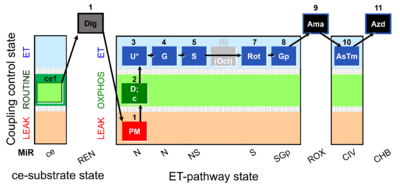 | D004_ce1;1Dig;1PM;2D;2c;3U;4G;5S;(6Oct);7Rot;8Gp;9Ama;10AsTm;11Azd | A - SUIT-001 - SUIT reference protocol Reference protocol RP1 for living PBMC and PLT cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-001 O2 mt D001 | O2 | 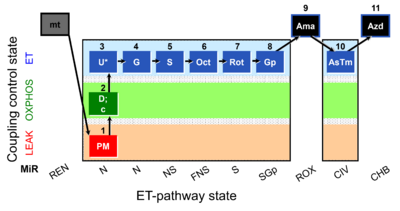 | D001_mt;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd | SUIT-001 - SUIT reference protocol Reference protocol RP1 for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-001 O2 pfi D002 | O2 | 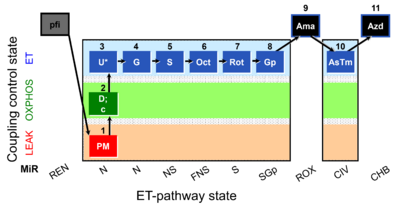 | D002_pfi;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd | SUIT-001 - SUIT reference protocol Reference protocol RP1 for permeabilized fibers |
| SUIT-002 |  | A: Doerrier 2018 Methods Mol Biol | ||
| SUIT-002 O2 ce-pce D007 | O2 | 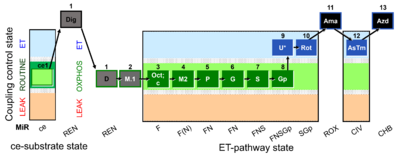 | D007_ce1;1Dig;1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for living cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-002 O2 ce-pce D007a | O2 |  | D007a_ce1;1Dig;1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for for living PBMC and PLT cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-002 O2 mt D005 | O2 | 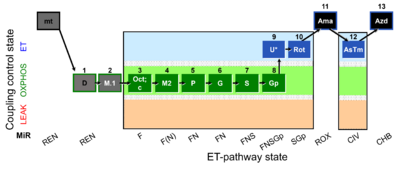 | D005_mt;1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-002 O2 pfi D006 | O2 | 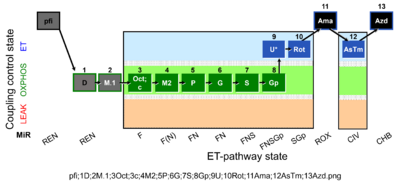 | D006_pfi;1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd | A -SUIT-002 - SUIT reference protocol Reference protocol RP2 for permeabilized fibers (pfi) |
| SUIT-003 |   | A: Coupling-control protocol, living cells »MiPNet08.09 CellRespiration Bioblast pdf | ||
| SUIT-003 AmR ce D017 | AmR | 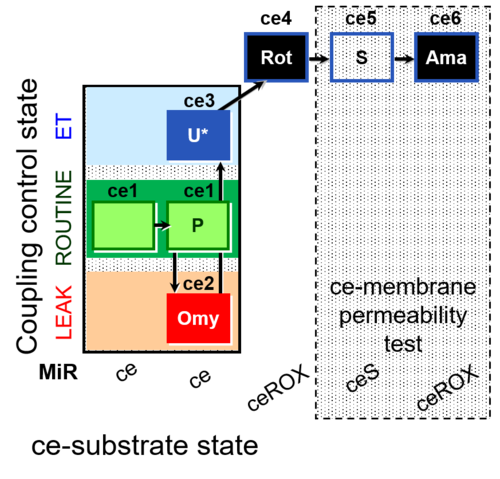 | D017_ce1;ce2P;ce3Omy;ce4U;ce5Rot;ce6S;ce7Ama | B: CCP(P) and cell membrane permeability test with succinate - SUIT-003 |
| SUIT-003 AmR ce D058 | AmR | 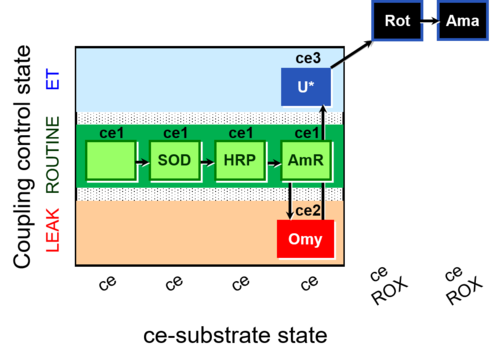 | D058_ce1;ce1SOD;ce1HRP;ce1AmR;ce2Omy;ce3U;ce4Rot;ce5Ama | B: Coupling control in intact cells with oligomycin to study the inhibitory effect of the AmR assay components on the respiration ; ce: non-permeabilized cells -SUIT-003 |
| SUIT-003 AmR ce D059 | AmR | 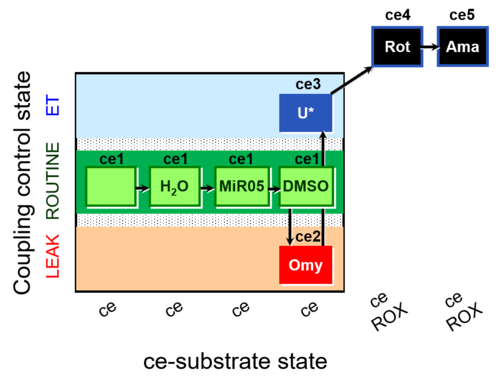 | D059_ce1;ce1H2O;ce1MiR05;ce1DMSO;ce2Omy;ce3U;ce4Rot;ce5Ama | B: Experimental control for SUIT-003 AmR ce D058 |
| SUIT-003 Ce1;ce1P;ce3U;ce4Glc;ce5M;ce6Rot;ce7S;1Dig;1c;2Ama;3AsTm;4Azd | O2 | 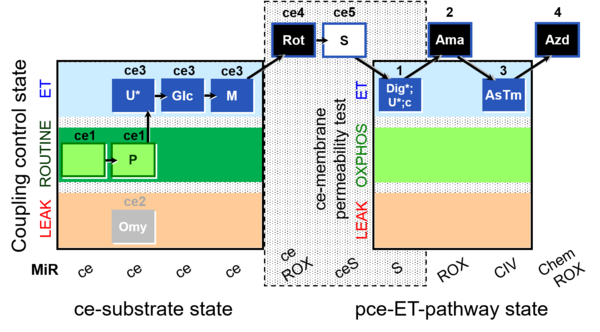 | C SUIT-003 Coupling-control protocol and respirometric cell viability test (CCV protocol) without oligomycin. | |
| SUIT-003 Ce1;ce1SD;ce2Omy;ce3U- | O2 | 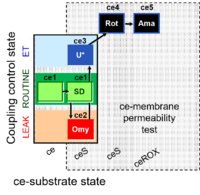 | B SUIT-003 Stadlmann 2006 Cell Biochem Biophys | |
| SUIT-003 Ce1;ce1SD;ce3U;ce4Rot;ce5Ama | O2 | 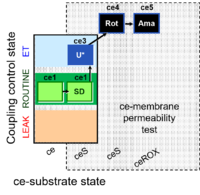 | B SUIT-003 CCP(S) | |
| SUIT-003 Ce1;ce2SD;ce3Omy;ce4U- | O2 | 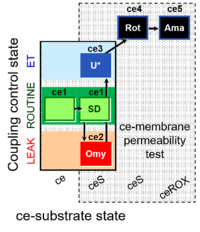 | B SUIT-003 Stadlmann 2006 Cell Biochem Biophys | |
| SUIT-003 Ce1;ce2SD;ce3U;ce4Rot;ce5Ama | O2 | 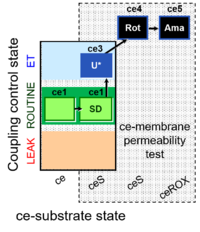 | B Steinlechner-Maran 1997 Transplantation | |
| SUIT-003 Ce1;ce2U;ce3Rot;ce4S;ce5Ama | O2 | 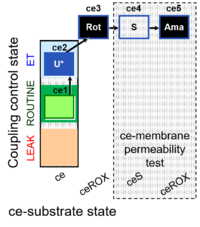 | B SUIT-003 CCP(S) | |
| SUIT-003 Ce1;ce3U;ce4Rot;ce5S;ce6Ama | O2 | 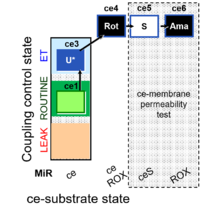 | B SUIT-003 CCP(S) | |
| SUIT-003 O2 ce D009 | O2 | 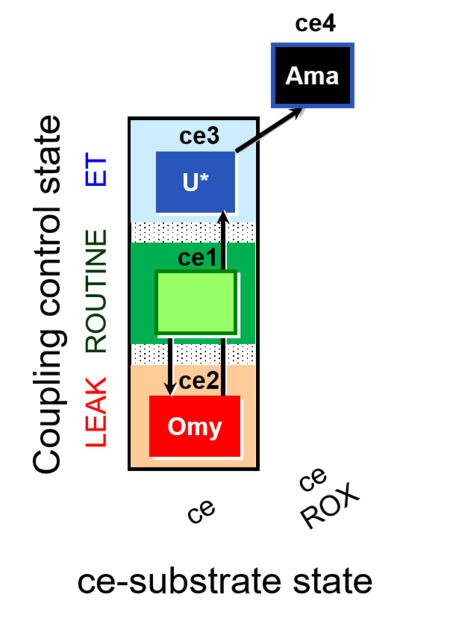 | D009_ce1;ce2Omy;ce3U;ce4Ama | A: - SUIT-003 - Coupling-control protocol (CCP) in living cells (ce) with oligomycin (LEAK state) |
| SUIT-003 O2 ce D012 | O2 | 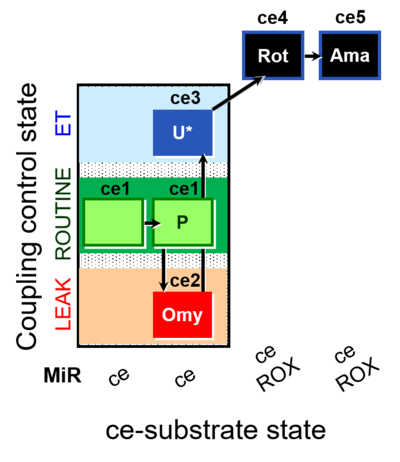 | D012_ce1;ce1P;ce2Omy;ce3U;ce4Rot;ce5Ama | B - SUIT-003 |
| SUIT-003 O2 ce D028 | O2 | 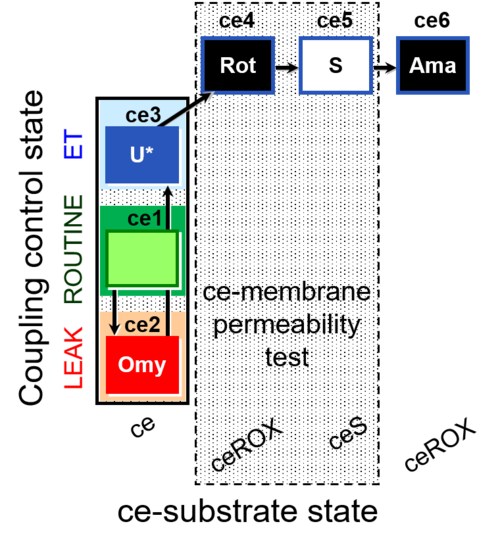 | D028_ce1;ce2Omy;ce3U;ce4Rot;ce5S;ce6Ama | B SUIT-003 Coupling-control protocol with succinate for permeability test |
| SUIT-003 O2 ce D037 | O2 | 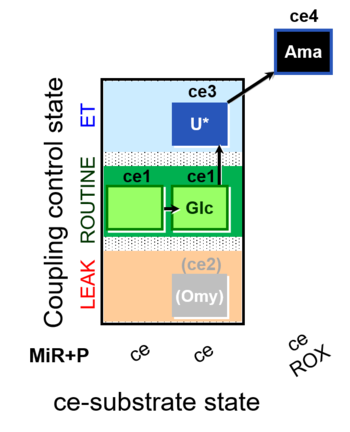 | D037_ce1;ce1Glc;(ce2Omy);ce3U;ce4Ama | A SUIT-003 - Coupling-control protocol and Crabtree effect |
| SUIT-003 O2 ce D038 | O2 | 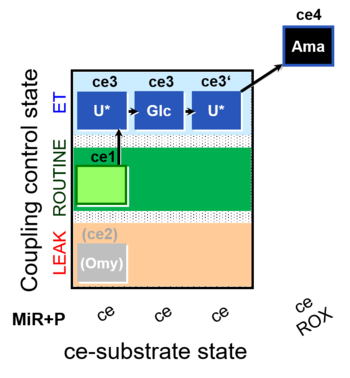 | D038_ce1;(ce2Omy);ce3U;ce3Glc;ce3'U;ce4Ama | A Coupling-control protocol and Crabtree effect test -SUIT-003 complex cell coupling-control protocol |
| SUIT-003 O2 ce D039 | O2 | 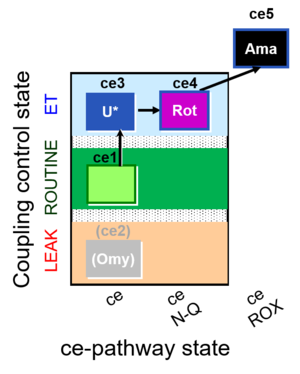 | D039_ce1;(ce2Omy);ce3U;ce4Rot;ce5Ama | A: Coupling control in intact cells without oligomycin; ce: non-permeabilized cells -SUIT-003 |
| SUIT-003 O2 ce D050 | O2 | 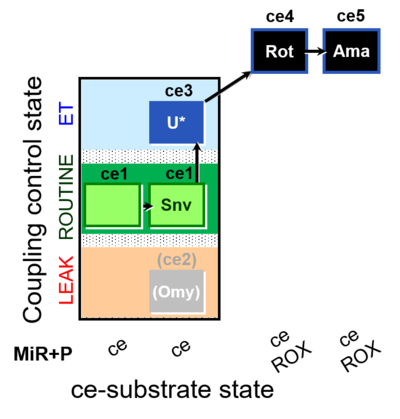 | D050_ce1;ce1Snv;(ce2Omy);ce3U;ce4Rot;ce5Ama | A: - Coupling-control protocol for living cells with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) - SUIT-003 - Plasma membrane-permeable succinate (NV118) in ROUTINE respiration. Coupling-control protocol (CCP) in living cells (ce) |
| SUIT-003 O2 ce D060 | O2 | 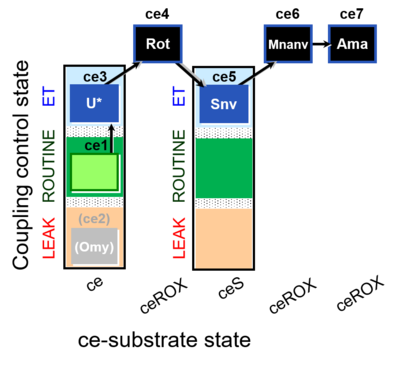 | D060_ce1;(ce2Omy);ce3U;ce4Rot;ce5Snv;ce6Mnanv;ce7Ama | B - coupling-control protocol for living cells with cell permeable compounds MitoKit-CII/Succinate-nv and MitoKit-CII/Malonate-nv -SUIT-003 - Coupling-control protocol (CCP) with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) and plasma membrane-permeable malonate (MitoKit-CII/Malonate-nv) in living cells (ce). |
| SUIT-003 O2 ce D061 | O2 | 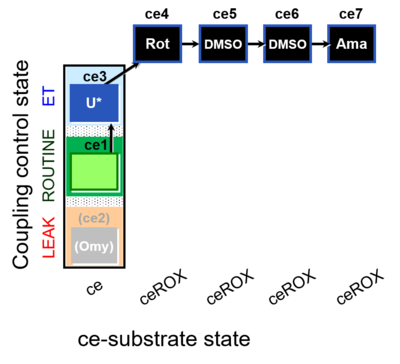 | D061_ce1;(ce2Omy);ce3U;ce4Rot;ce5DMSO;ce6DMSO;ce7Ama | B Carrier control for the coupling-control protocol for living cells with cell permeable compounds MitoKit-CII/Succinate-nv and MitoKit-CII/Malonate-nv - SUIT-003 O2 ce D060 |
| SUIT-003 O2 ce D062 | O2 | 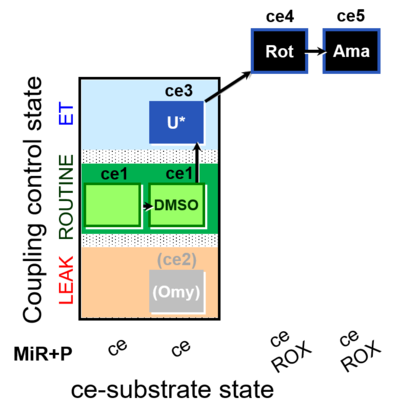 | D062_ce1;ce1DMSO;(ce2Omy);ce3U;ce4Rot;ce5Ama | A: Carrier control for the Coupling-control protocol for living cells with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) - SUIT-003 O2 ce D050 |
| SUIT-003 O2 ce-pce D013 | O2 | 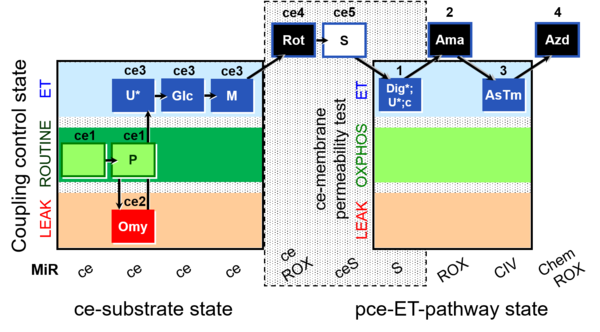 | D013_ce1;ce1P;ce2Omy;ce3U;ce3Glc;ce3M;ce4Rot;ce5S;1Dig;1U;1c;2Ama;3AsTm;4Azd | C - SUIT-003 |
| SUIT-003 O2 ce-pce D018 | O2 | 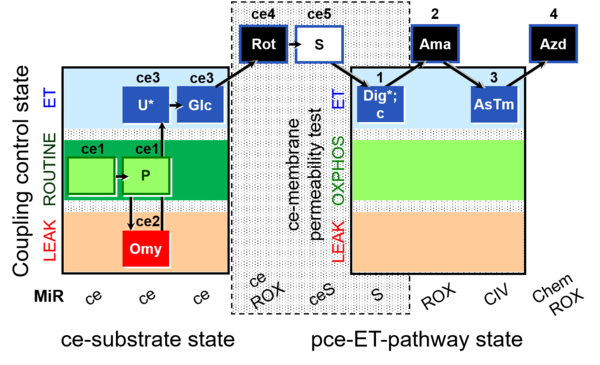 | D018_ce1;ce1P;ce2Omy;ce3U;ce3Glc;ce4Rot;ce5S;1Dig;1c;2Ama;3AsTm;4Azd | C SUIT-003 Second version of the Coupling-control protocol and respirometric cell viability test (CCV protocol) |
| SUIT-003 O2 ce-pce D020 | O2 | 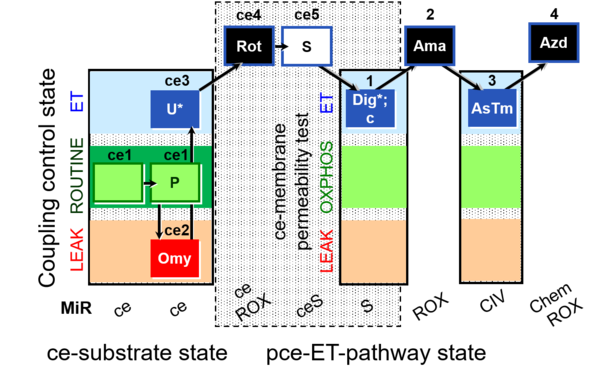 | D020_ce1;ce1P;ce2Omy;ce3U;ce4Rot;ce5S;1Dig;1c;2Ama;3AsTm;4Azd | A SUIT-003 - Coupling-control protocol and respirometric cell viability test; ce: living cells, pce: permeabilized cells |
| SUIT-003 pH ce D067 | O2 | 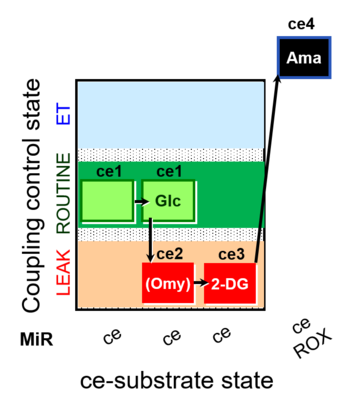 | D067_ce1;ce1Glc;ce2Omy;ce3DG;ce4Ama | A Coupling-control protocol and Crabtree effect with glycolysis inhibition- SUIT-003 |
| SUIT-004 |  | |||
| SUIT-004 O2 pfi D010 | O2 | 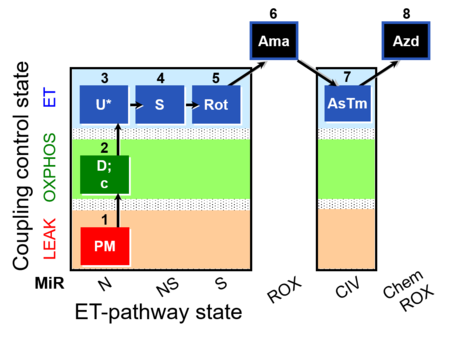 | D010_1PM;2D;2c;3U;4S;5Rot;6Ama;7AsTm;8Azd | SUIT-004 - Short protocol linked to SUIT-001 O2 pfi D002 SUIT RP1 (human skeletal muscle) |
| SUIT-005 |  | A: when malate-anaplerotic activity is zero | ||
| SUIT-005 O2 pfi D011 | O2 | 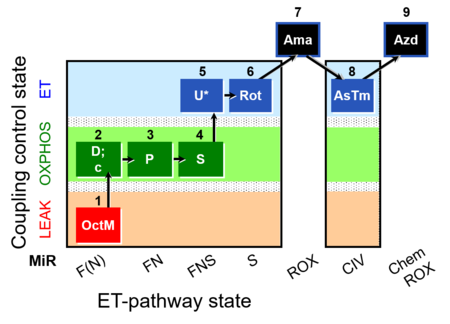 | D011_1OctM;2D;2c;3P;4S;5U;6Rot;7Ama;8AsTm;9Azd | A: - SUIT-005 short protocol linked to SUIT-002 O2 pfi D006 - SUIT RP2 (human skeletal muscle) |
| SUIT-006 |  | A: Coupling-control protocol, mtprep | ||
| SUIT-006 AmR mt D048 | AmR | 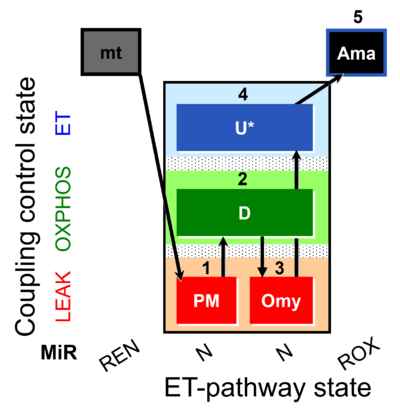 | D048_1PM;2D;3Omy;4U;5Ama | A: short protocol for simultaneous determination of O2 flux and H2O2 in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells)- SUIT-006 |
| SUIT-006 Fluo mt D034 | Fluo | 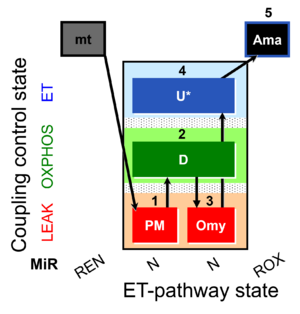 | D034_1PM;2D;3Omy;4U;5Ama | A: short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) -SUIT-006 |
| SUIT-006 MgG ce-pce D085 | MgG | 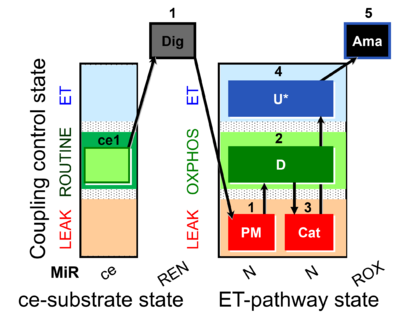 | D085_ce1;1Dig;1PM;2D;3Cat;4U;5Ama | A: short coupling-control protocol for simultaneous determination of O2 flux and mitochondrial ATP production in permeabilized cells -SUIT-006 |
| SUIT-006 MgG mt D055 | MgG | 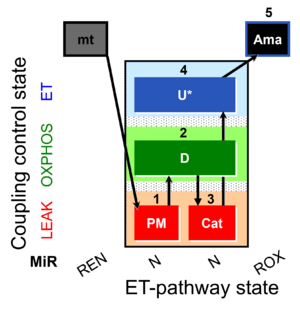 | D055_1PM;2D;3Cat;4U;5Ama | A: short coupling-control protocol for simultaneous determination of O2 flux and mitochondrial ATP production in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) -SUIT-006 |
| SUIT-006 O2 ce-pce D029 | O2 | 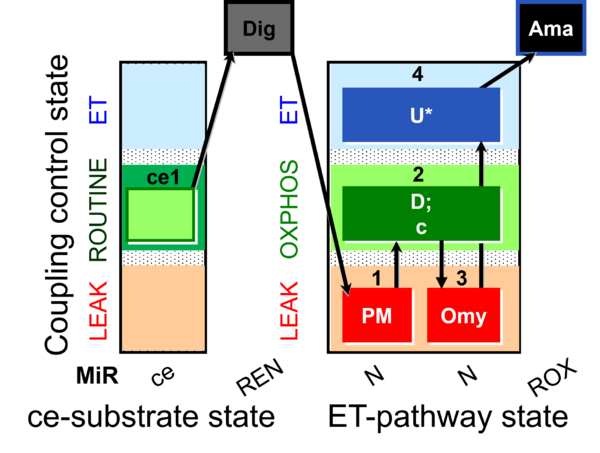 | D029_ce1;1Dig;1PM;2D;2c;3Omy;4U;5Ama | A to analyse the coupling control on permeabilized cells in N(PM) pathway control state- SUIT-006 |
| SUIT-006 O2 mt D022 | O2 | 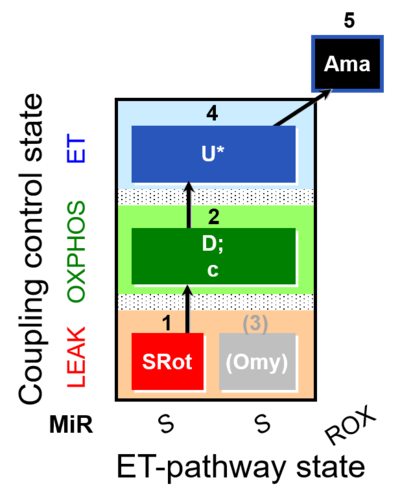 | D022_1SRot;2D;2c;(3Omy);4U;5Ama | A - SUIT-006 - Protocol for coupling control assessment in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) |
| SUIT-006 O2 mt D047 | O2 | 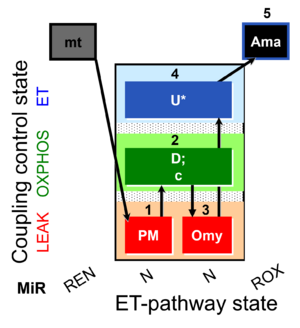 | D047_1PM;2D;2c;3Omy;4U;5Ama | A SUIT-006 - is a coupling-control protocol in the N-pathway for isolated mitochondria, tissue homogenate and cells permeabilized before addition to the O2k chamber. |
| SUIT-006 Q ce-pce D073 | Q | 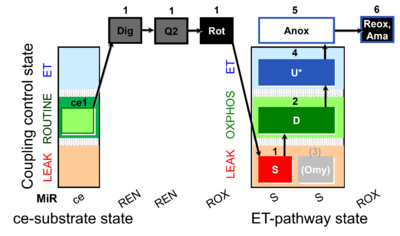 | D073_ce1;1Dig;1Q2;1Rot;1S;2D;(3Omy);4U;5Anox;6Ama.png | A Coupling control protocol to analyze mitochondrial O2 consumption and the redox state of the Q-pool in S(Rot)-pathway control state in permeabilized cells- SUIT-006 |
| SUIT-006 Q mt D071 | Q | 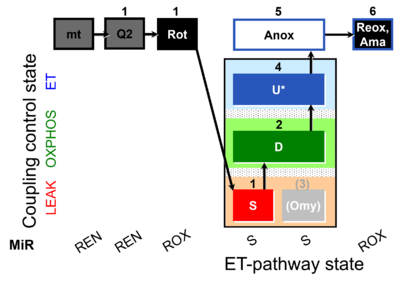 | D071_mt;1Q2;1Rot;1S;2D;(3Omy);4U;5Anox;6Ama.png | A Coupling-control protocol to analyze mitochondrial O2 consumption and the redox state of the Q-pool in S(Rot)-pathway control state in isolated mitochondria- SUIT-006 |
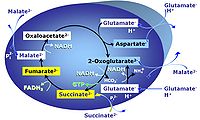
| SUIT-007 |  | A: Glutamate anaplerotic pathway | ||
| SUIT-007 O2 ce-pce D030 | O2 | 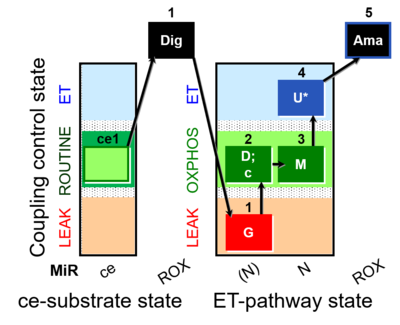 | D030_ce1;1Dig;1G;2D;2c;3M;4U;5Ama | A pce: permeabilized cells - SUIT-007 |
| SUIT-008 |  | A: Additivity between the N- and S-pathway in the Q-junction | ||
| SUIT-008 O2 ce-pce D025 | O2 | 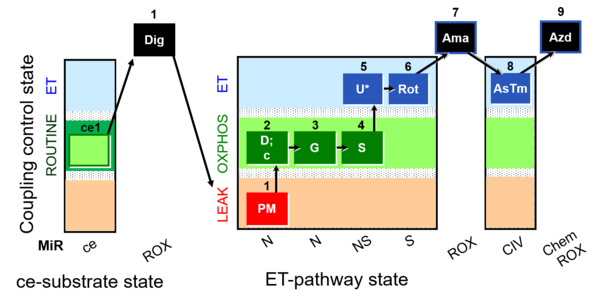 | D025_ce1;1Dig;1PM;2D;2c;3G;4S;5U;6Rot;7Ama;8AsTm;9Azd | A: for cells-permeabilized cells Lemieux 2017 Sci Rep - SUIT-008 |
| SUIT-008 O2 mt D026 | O2 | 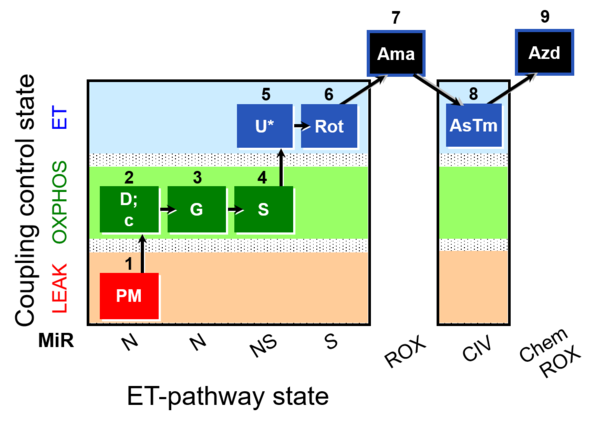 | D026_1PM;2D;2c;3G;4S;4D;5U;6Rot;7Ama;8AsTm;9Azd | A: SUIT protocol for mt; mt: isolated mitochondria and tissue homogenate - SUIT-008 |
| SUIT-008 O2 pce D25 | O2 | 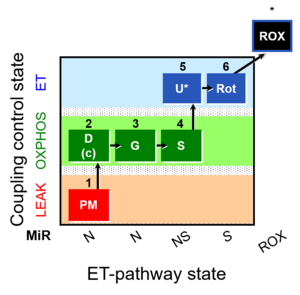 | C25_1PM;2D;2c;3G;4S;4D;5U;6Rot;7Ama;8AsTm;9Azd | B: SUIT 8 for permeabilized cells Lemieux 2017 Sci Rep |
| SUIT-008 O2 pfi D014 | O2 | 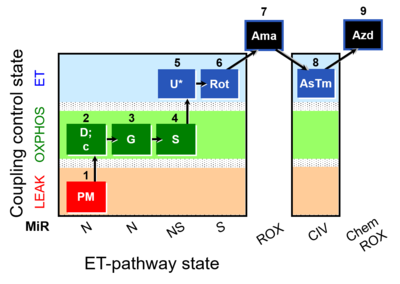 | D014_1PM;2D;2c;3G;4S;4D;5U;6Rot;7Ama;8AsTm;9Azd | A - Lemieux 2017 Sci Rep - SUIT-008- O2 pfi D014 protocol was used a a reference protocol in MitoEAGLE project (WG2) to evaluate the researcher skills preparing permeabilized fibers |
| SUIT-009 AmR ce-pce D019 | 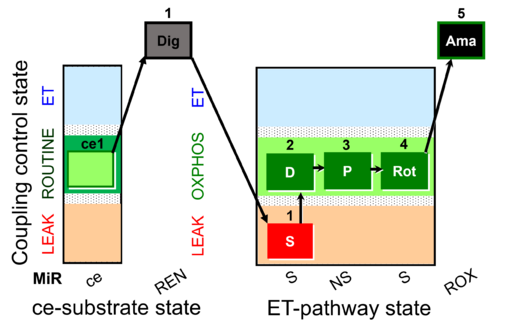 | D019_ce1;1Dig;1S;2D;3P;4Rot;5Ama | B: short protocol for H2O2 (AmR) in ce: non-permeabilized cells, pce: permeabilized cells -SUIT-009 | |
| SUIT-009 AmR mt D021 | AmR | 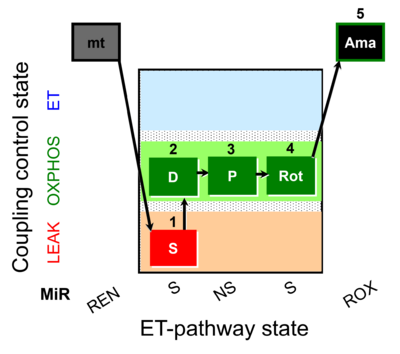 | D021_1S;2D;3P;4Rot;5Ama | B: short protocol for H2O2 (AmR) in mitochondrial preparations; mt: isolated mitochondria, tissue homogenate and permeabilized cells - SUIT-009short protocol for H2O2 (AmR) for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-009 O2 ce-pce D016 | 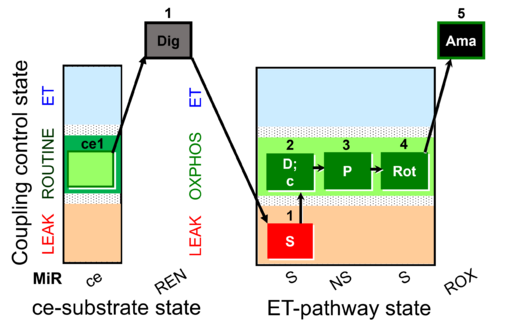 | D016_ce1;1Dig;1S;2D;3P;4Rot;5Ama | B - SUIT-009 - short protocol for mt-respiration in ce: non-permeabilized cells, pce: permeabilized cells. | |
| SUIT-009 O2 mt D015 | 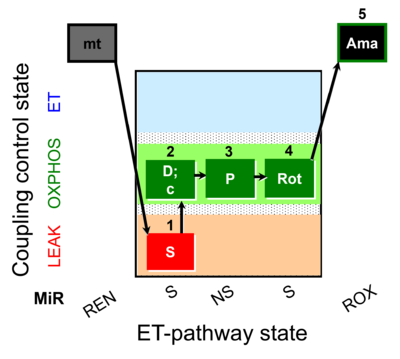 | D015_1S;2D;3P;4Rot;5Ama | B - SUIT-009- short protocol for respiration in mitochondrial preparations; mt: isolated mitochondria, tissue homogenate and permeabilized cells | |
| SUIT-010 |  | A: Optimization of digitonin concentration for pce | ||
| SUIT-010 O2 ce-pce D008 | O2 |  | D008_ce1;ce2Rot;ce3S;ce4D;1Dig;1c | A -SUIT-010 - Optimum digitonin concentration for pce |
| SUIT-011 |  | A: Maximum mitochondrial respiratory capacity (OXPHOS with NS substrates) and coupling/pathway control | ||
| SUIT-011 O2 pfi D024 | O2 | 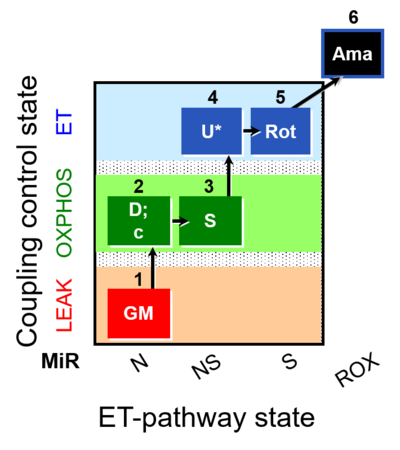 | D024_1GM;2D;2c;3S;4U;5Rot;6Ama | A pfi: permeabilized fibers - SUIT-011 |
| SUIT-012 |  | A: Coupling control (L- P- E) with NADH-linked substrates (PM and PGM) | ||
| SUIT-012 O2 ce-pce D052 | O2 | 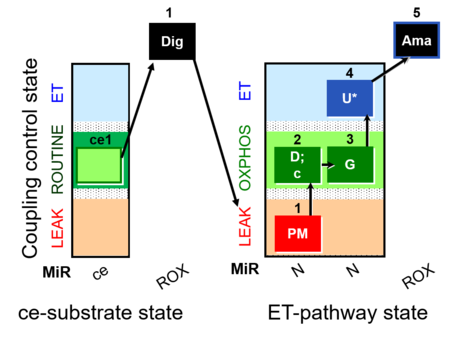 | D052_ce1;1Dig;1PM;2D;2c;3G;4U;5Ama | A Linear coupling control with NADH-linked substrates for pce - SUIT-012 |
| SUIT-012 O2 mt D027 | O2 | 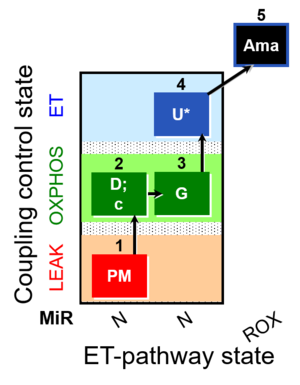 | D027_1PM;2D;2c;3G;4U;5Ama | A Linear coupling control with NADH-linked substrates for isolated mitochondria, tissue homogenate and permeabilized cells- SUIT-012 |
| SUIT-013 |  | A | ||
| SUIT-013 AmR ce D023 | AmR | 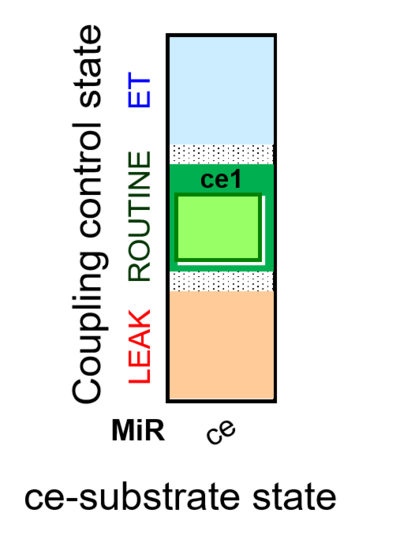 | D023_ce1 | A SUIT-013 |
| SUIT-014 |  | A: Cells or tissue types that display a preference for GM over PM to support NADH-linked respiration | ||
| SUIT-014 O2 pfi D042 | O2 | 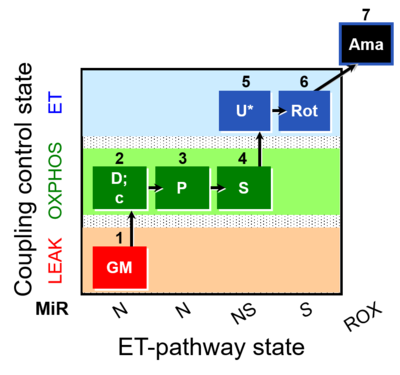 | D042_1GM;2D;2c;3P;4S;5U;6Rot;7Ama | A SUIT-014 |
| SUIT-015 |  | A: F-pathway in LEAK state and OXPHOS state | ||
| SUIT-015 O2 pti D043 | O2 | 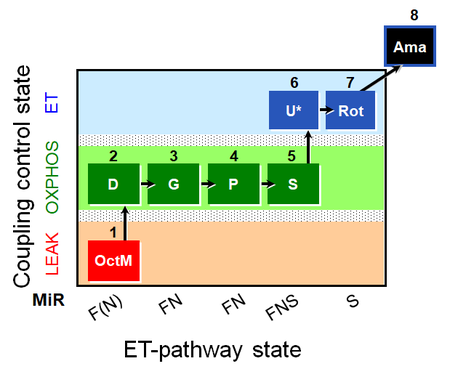 | D043_1OctM;2D;2c;3G;4P;5S;6U;7Rot;8Ama | A pti: permeabilized tissue- SUIT-015 |
| SUIT-016 |  | A: F-pathway in LEAK state and OXPHOS state | ||
| SUIT-016 O2 pfi D044 | O2 | 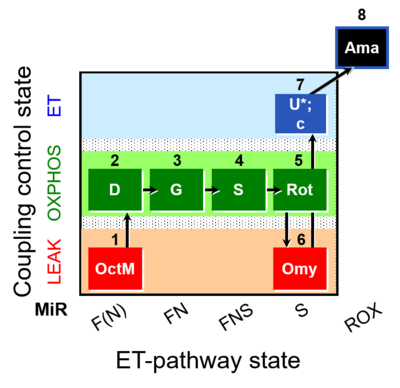 | D044_1OctM;2D;3G;4S;5Rot;6Omy;7U;8Ama | A pfi: permeabilized fibers- SUIT-016 |
| SUIT-017 |  | A: | ||
| SUIT-017 O2 mt D046 | O2 | 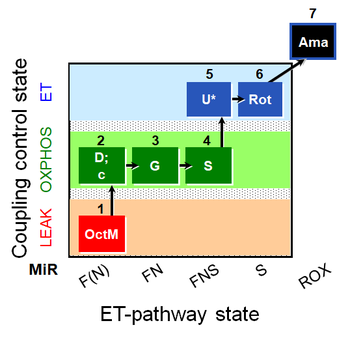 | D046_1OctM;2D;2c;3G;3c;4S;5U;6Rot;7Ama | A: SUIT-017 |
| SUIT-017 O2 pfi D049 | O2 |  | D049_1OctM;2D;2c;3G;4S;5U;6Rot;7Ama | A: SUIT-017 |
| SUIT-018 |  | A: A protocol for oxygen dependence of O2 flux and H2O2 flux in isolated mitochondria,tissue homogenate or permeabilized cells | ||
| SUIT-018 AmR ce-pce D068 | AmR | 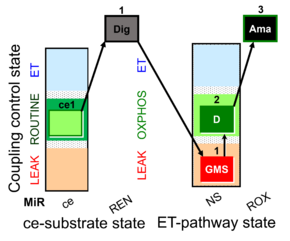 | D068_ce1;1Dig;1GMS;2D;3Ama | A -SUIT-018 - SUIT protocol for simultaneous determination of O2 and H2O2 flux in permeabilized cells at various oxygen concentrations |
| SUIT-018 AmR mt D031 | AmR | 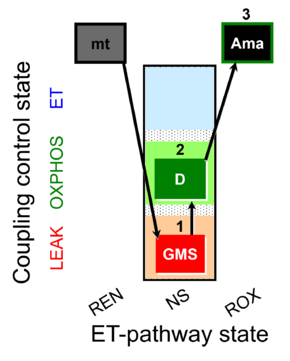 | D031_mt;1GMS;2D;3Ama | A -SUIT-018 - SUIT protocol for simultaneous determination of O2 and H2O2 flux in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) at low oxygen concentration (tissue normoxia) |
| SUIT-018 AmR mt D040 | AmR |  | D040_mt;1GMS;2D;3Ama | A: simultaneous determination of O2 and H2O2 flux in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells)-SUIT-018 |
| SUIT-018 AmR mt D041 | AmR |  | D041_mt;1GMS;2D;3Ama | A -SUIT-018 |
| SUIT-018 O2 mt D054 | O2 | 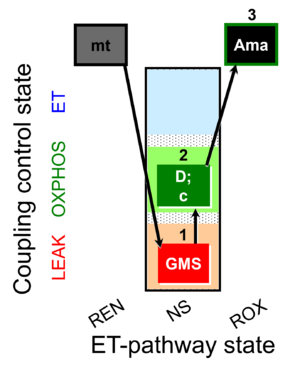 | D054_mt;1GMS;2D;2c;3Ama | B: SUIT-018 |
| SUIT-019 |  | A: | ||
| SUIT-019 O2 pfi D045 | O2 | 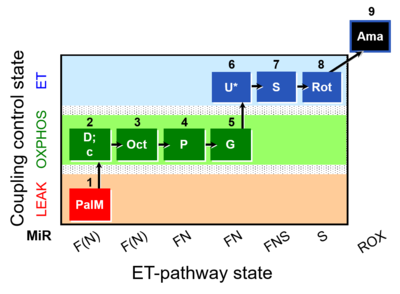 | D045_1PalM;2D;2c;3Oct;4P;5G;6U;7S;8Rot;9Ama | A SUIT-019 |
| SUIT-020 |  | A: simultaneous determination of O2 flux and mt-membrane potential | ||
| SUIT-020 Fluo mt D033 | Fluo | 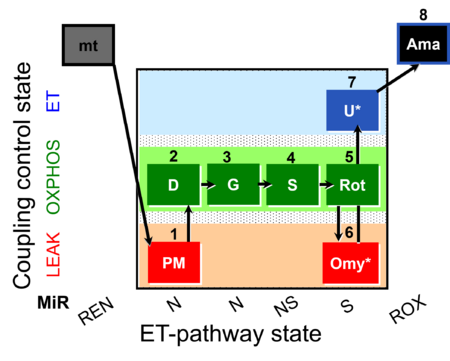 | D033_1PM;2D;3G;4S;5Rot;6Omy;7U;8Ama | A:short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)- SUIT-020 |
| SUIT-020 O2 mt D032 | O2 | 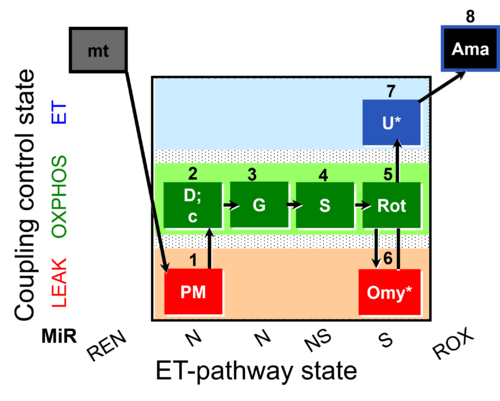 | D032_1PM;2D;2c;3G;4S;5Rot;6Omy;7U;8Ama | A: short protocol for determination of O2 flux in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)-SUIT-020 |
| SUIT-021 |  | A: simultaneous determination of O2 flux and mt-membrane potential | ||
| SUIT-021 Fluo mt D036 | Fluo | 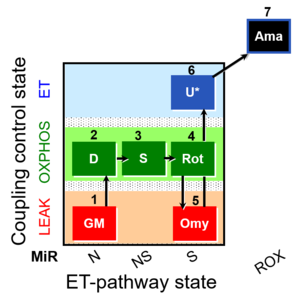 | D036_1GM;2D;3S;4Rot;5Omy;6U;7Ama | A: short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)-SUIT-021 |
| SUIT-021 O2 mt D035 | O2 | 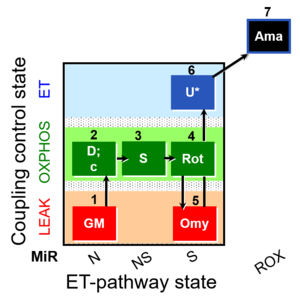 | D035_1GM;2D;2c;3S;4Rot;5Omy;6U;7Ama | A:short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permabilized cells, and tissue homogenate)-SUIT-021 |
| SUIT-022 |  | A: Determination of the respiration due to the alternative oxidase pathway | ||
| SUIT-022 O2 ce D051 | O2 | 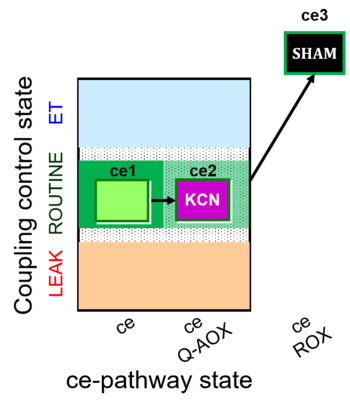 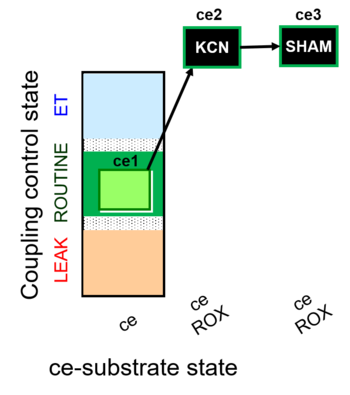 | D051_ce1;ce2KCN;ce3SHAM | A Determination of the respiration due to the alternative oxidase pathway -SUIT-022 |
| SUIT-023 |  | A: Determination of respiration through the CIII-CIV pathway | ||
| SUIT-023 O2 ce D053 | O2 | 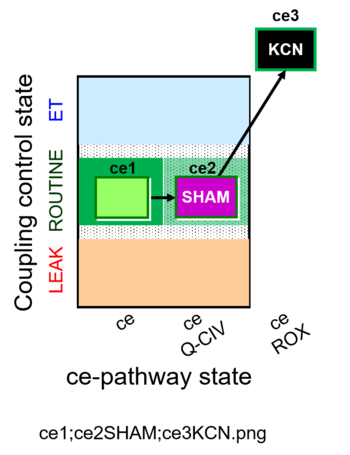 | D053_ce1;ce2SHAM;ce3KCN | A Determination of the respiration due to the cytochrome c oxidase pathway -SUIT-023 |
| SUIT-024 |  | A: Determination of ATPase activity in mitochondrial preparations. | ||
| SUIT-024 O2 ce-pce D056 | O2 | 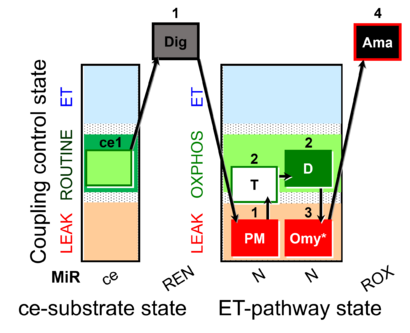 | D056_ce1;1Dig;1PM;2T;2D;3Omy;4Ama | Determination of the presence of ATPases in mitochondrial preparations - SUIT-024 |
| SUIT-025 |  | |||
| SUIT-025 O2 mt D057 | O2 | 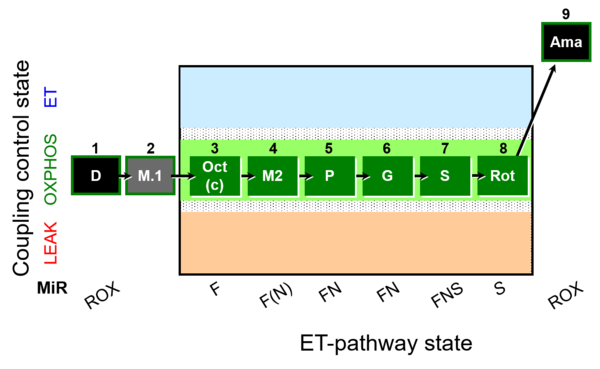 | D057_1D;2M;3Oct;3c;4M;5P;6G;7S;8Rot;9Ama | - SUIT-025 |
| SUIT-026 |  | A Short protocol to study RET-related H2O2 production | ||
| SUIT-026 AmR ce-pce D087 | AmR | 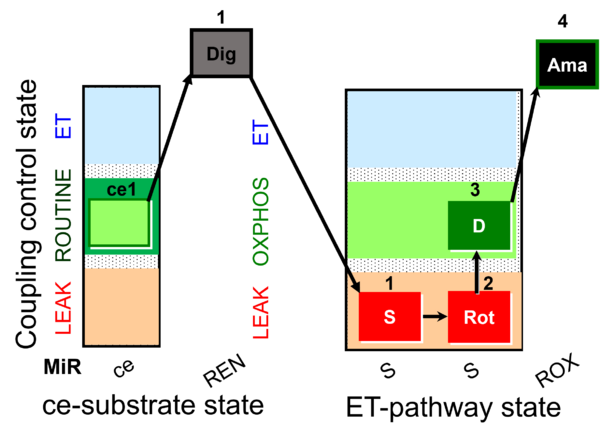 | D087_ce1;1Dig;1S;2Rot;3D;4Ama | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in permeabilized cells |
| SUIT-026 AmR mt D064 | AmR | 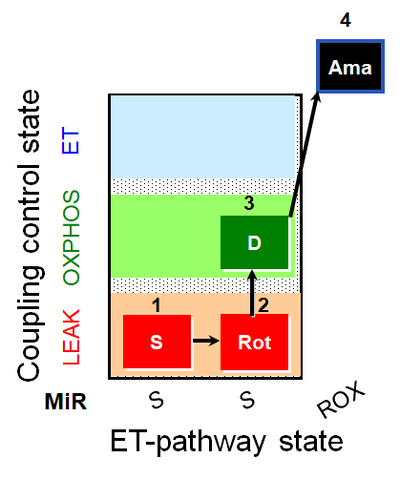 | D064_1S;2Rot;3D;4Ama | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in mtprep |
| SUIT-026 AmR mt D077 | AmR |  | D077_1S;2Rot;3D;4Ama | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in mtprep |
| SUIT-026 O2 ce-pce D088 | O2 | 400px | D088_ce1;1Dig;1S;2Rot;3D;3c;4Ama | B - SUIT-026 - Protocol for the respiratory control of SUIT-026 AmR ce-pce D087 |
| SUIT-026 O2 mt D063 | O2 | 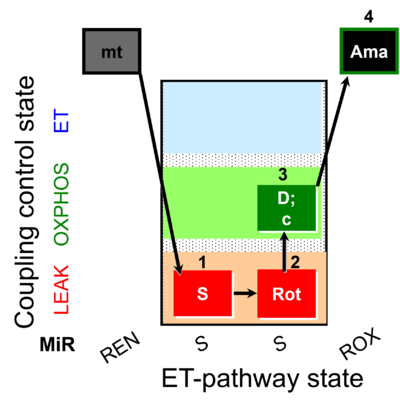 | D063_1S;2Rot;3D;3c;4Ama | B - SUIT-026 - Protocol for the respiratory control of SUIT-026 AmR mt D064 |
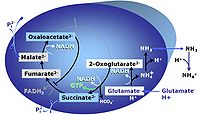
| SUIT-027 |  | A: Malate anaplerotic pathway | ||
| SUIT-027 O2 ce-pce D065 | O2 | 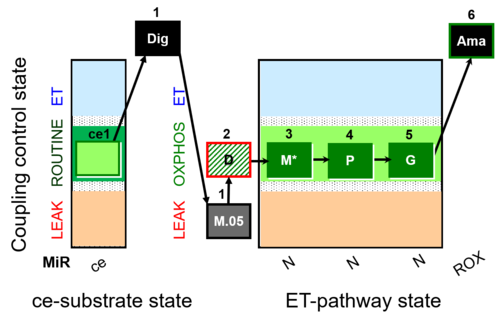 | D065_ce1;1Dig;1M;2D;3M;4P;5G | A - SUIT-027 - Protocol for analysis of the malate anaplerotic pathway |
| SUIT-028 | 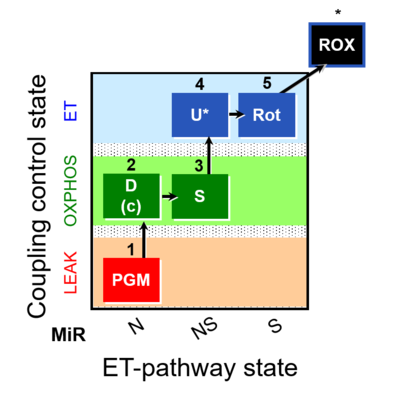 | C MiPNet18.13 IOC84 Alaska | ||
| SUIT-029 O2 mt D066 | O2 | 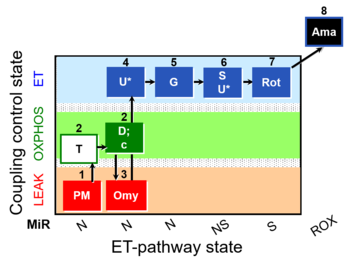 | D066_mt;1PM;2T;2D;2c;3Omy;4U;5G;6S;6U;7Rot;8Ama | A: quality control evaluation of mitochondrial preparations - SUIT-029 |
| SUIT-031 | 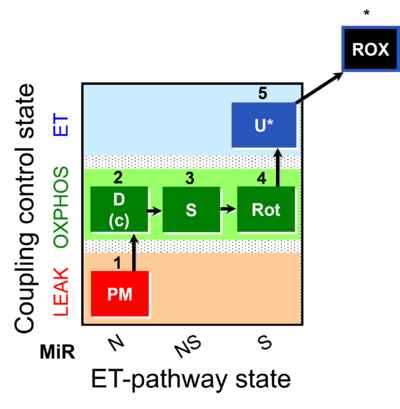 | A: | ||
| SUIT-031 O2 ce-pce D079 | O2 | 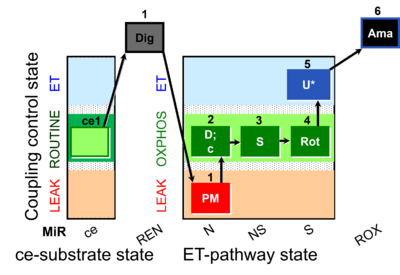 | D079_ce1;1Dig;1PM;2D;2c;3S;4Rot;5U;6Ama | A to analyse O2 flux in the NS- pathway control state in permeabilized cells- SUIT-031 |
| SUIT-031 O2 mt D075 | O2 | 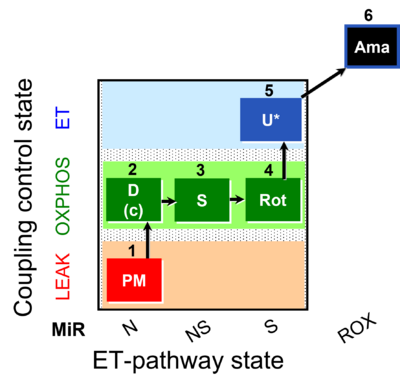 | D075_1mt;1PM;2D;2c;3S;4Rot;5U;6Ama | A to analyse O2 flux in the NS- pathway control state on isolated mitochondria- SUIT-031 |
| SUIT-031 Q ce-pce D074 | Q | 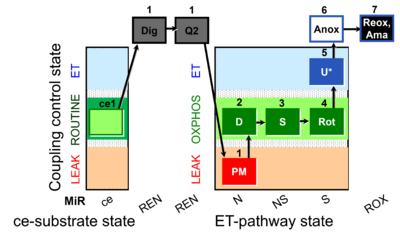 | D074_ce1;1Dig;1Q2;1PM;2D;3S;4Rot;5U;6Anoxia;7Ama | A to analyse O2 flux and the Q-redox state in the NS- pathway control state on permeabilized cells- SUIT-031 |
| SUIT-031 Q mt D072 | Q | 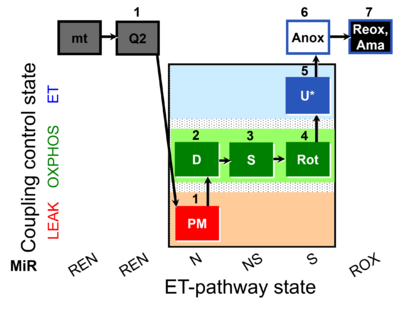 | D072_mt;1Q2;1PM;2D;3S;4Rot;5U;6Anox;7Ama | A to analyse O2 flux and the Q-redox state in the N-,NS- and S- pathway control state on isolated mitochondria- SUIT-031 |
| SUITbrowser | Use the SUITbrowser to find the substrate-uncoupler-inhibitor-titration (SUIT) protocol most suitable for addressing your research questions.
Open the SUITbrowser: http://suitbrowser.oroboros.at/
 How to find a DL-Protocol (DLP) How to find a DL-Protocol (DLP) |
Glossary
- » List of SUIT states
List of SUIT states
| Term | Abbreviation | Description |
|---|---|---|
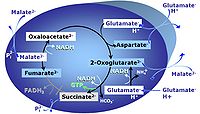
| Anaplerotic pathway control state | a | Anaplerotic pathway control states are fuelled by single substrates which are transported into the mitochondrial matrix and increase the pool of intermediates of the tricarboxylic acid cycle. Malic enzyme (mtME), phosphoenopyruvate carboxykinase (PEPCK), propionyl-CoA carboxylase, and pyruvate carboxylase play important roles in anaplerosis. The glutamate-anaplerotic pathway control state and malate-anaplerotic pathway control state are the most important anaplerotic substrate control states (aN). |
| Complex I&II-linked substrate state | NS | See NS-pathway control state (previous: CI&II-linked) |
| Complex I-linked substrate state | N | See N-pathway control state (previous: CI-linked) versus Complex I |
| Complex II-linked substrate state | SRot, S | See S-pathway control state (previous: CII-linked) |
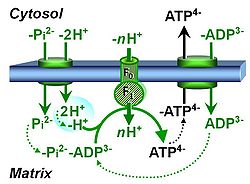
| Coupling-control state | CCS | Coupling-control states are defined in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, homogenates) as LEAK respiration, OXPHOS, and ET states, with corresponding respiration rates (L, P, E) in any electron-transfer-pathway state which is competent for electron transfer. These coupling states are induced by titration of ADP and uncouplers, and application of specific inhibitors of the phosphorylation pathway. In living cells, the coupling-control states are LEAK respiration, ROUTINE, and ET states of respiration with corresponding rates L, R, E, using membrane-permeable inhibitors of the phosphorylation system (e.g. oligomycin) and uncouplers (e.g. CCCP). Coupling-control protocols induce these coupling-control states sequentially at a constant electron-transfer-pathway state. |
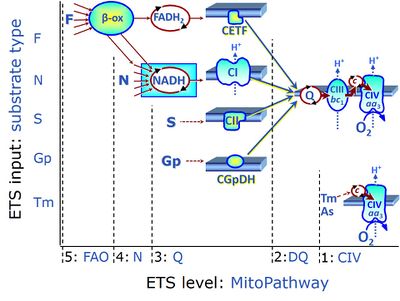
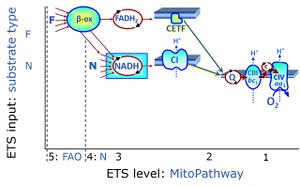
| Electron-transfer-pathway state | ET-pathway state |
Electron-transfer-pathway states are obtained in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, tissue homogenate) by depletion of endogenous substrates and addition to the mitochondrial respiration medium of fuel substrates (CHNO) activating specific mitochondrial pathways, and possibly inhibitors of specific pathways. Mitochondrial electron-transfer-pathway states have to be defined complementary to mitochondrial coupling-control states. Coupling-control states require ET-pathway competent states, including oxygen supply. Categories of SUIT protocols are defined according to mitochondrial ET-pathway states. » MiPNet article |
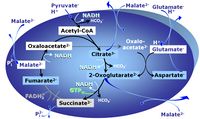
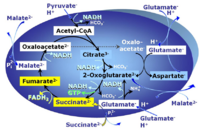
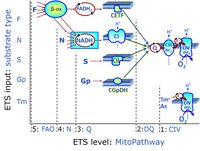
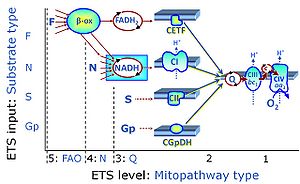
| FN | FN | FN is induced in mt-preparations by addition of NADH-generating substrates (N-pathway control state, or CI-linked pathway control) in combination with one or several fatty acids, which are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). This physiological substrate combination is required for partial reconstitution of TCA cycle function and convergent electron-input into the Q-junction, to compensate for metabolite depletion into the incubation medium. FS in combination exerts an additive effect of convergent electron flow in most types of mitochondria. |
| FNS | FNS | FNS is induced in mt-preparations by addition of NADH-generating substrates (N-pathway control state, or CI-linked pathway control) in combination with succinate (S-pathway control state; S- or CII-linked) and one or several fatty acids, which are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). This physiological substrate combination is required for partial reconstitution of TCA cycle function and convergent electron-input into the Q-junction, to compensate for metabolite depletion into the incubation medium. FNS in combination exerts an additive effect of convergent electron flow in most types of mitochondria. |
| FNSGp | FNSGp |
MitoPathway control state: FNSGp
SUIT protocol: SUIT-002 This substrate combination supports convergent electron flow to the Q-junction. |
| Fatty acid oxidation pathway control state | F, FAO | In the fatty acid oxidation pathway control state (F- or FAO-pathway), one or several fatty acids are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting relative to the N-pathway branch), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the type N substrate malate is required at low concentration (0.1 mM) as a co-substrate for FAO in mt-preparations, since accumulation of Acetyl-CoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of Acetyl-CoA into the TCA cycle catalyzed by citrate synthase. Peroxysomal β-oxidation carries out few β-oxidation cycles, thus shortening very-long-chain fatty acids (>C20) for entry into mitochondrial β-oxidation. Oxygen consumption by peroxisomal acyl-CoA oxidase is considered as residual oxygen consumption rather than cell respiration. |
| GM-pathway control state | GM | GM: Glutamate & Malate.
MitoPathway control state: NADH electron transfer-pathway state The GM-pathway control state (glutamate-malate pathway control state) is established when glutamate&malate are added to isolated mitochondria, permeabilized cells and other mitochondrial preparations. Glutamate and transaminase are responsible for the metabolism of oxaloacetate, comparable to the metabolism with acetyl-CoA and citrate synthase. |
| GMS-pathway control state | GMS | GMS: Glutamate & Malate & Succinate.
MitoPathway control: NS Transaminase catalyzes the reaction from oxaloacetate to 2-oxoglutarate, which then establishes a cycle without generation of citrate. OXPHOS is higher with GS (CI&II) compared to GM (CI) or SRot (CII). This documents an additive effect of convergent CI&II electron flow to the Q-junction, with consistent results obtained with permeabilized muscle fibres and isolated mitochondria (Gnaiger 2009). |
| Glutamate-anaplerotic pathway control state | G | G: Glutamate is an anaplerotic NADH-linked type 4 substrate (N). When supplied as the sole fuel substrate in the glutamate-anaplerotic pathway control state, G is transported by the electroneutral glutamate-/OH- exchanger, and is oxidised via mt-glutamate dehydrogenase in the mitochondrial matrix. The G-pathway plays an important role in glutaminolysis. |
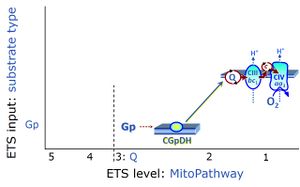
| Glycerophosphate pathway control state | Gp | The glycerophosphate pathway control state (Gp) is an ET-pathway level 3 control state, supported by the fuel substrate glycerophosphate and electron transfer through glycerophosphate dehydrogenase Complex into the Q-junction. The glycerolphosphate shuttle represents an important pathway, particularly in liver and blood cells, of making cytoplasmic NADH available for mitochondrial oxidative phosphorylation. Cytoplasmic NADH reacts with dihydroxyacetone phosphate catalyzed by cytoplasmic glycerophos-phate dehydrogenase. On the outer face of the inner mitochondrial membrane, mitochondrial glycerophosphate dehydrogenase oxidises glycerophosphate back to dihydroxyacetone phosphate, a reaction not generating NADH but reducing a flavin prosthesic group. The reduced flavoprotein donates its reducing equivalents to the electron transfer-pathway at the level of CoQ. |
| LEAK respiration | L | |
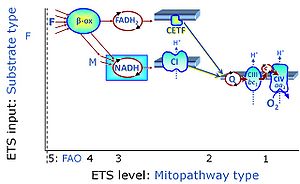
| Malate-anaplerotic pathway control state | M | M: Malate alone does not support respiration of mt-preparations if oxaloacetate cannot be metabolized further in the absence of a source of acetyl-CoA. Transport of oxaloacetate across the inner mt-membrane is restricted particularly in liver. Mitochondrial citrate and 2-oxoglutarate (α-ketoglutarate) are depleted by antiport with malate. Succinate is lost from the mitochondria through the dicarboxylate carrier. OXPHOS capacity with malate alone is only 1.3% of that with Pyruvate&Malate in isolated rat skeletal muscle mitochondria. However, many mammalian and non-mammalian mitochondria have a mt-isoform of NADP+- or NAD(P)+-dependent malic enzyme (mtME), the latter being particularly active in proliferating cells. Then the anaplerotic pathway control state with malate alone (aN) supports high respiratory activities comparable to the NADH-linked pathway control states (N) with pyruvate&malate or glutamate&malate substrate combinations (PM-pathway control state, GM-pathway control state). |
| NADH electron transfer-pathway state | N | The NADH electron transfer-pathway state (N) is obtained by addition of NADH-linked substrates (CI-linked), feeding electrons into the N-junction catalyzed by various mt-dehydrogenases. N-supported flux is induced in mt-preparations by the addition of NADH-generating substrate combinations of pyruvate (P), glutamate (G), malate (M), oxaloacetate (Oa), oxoglutarate (Og), citrate, hydroxybutyrate. These N-junction substrates are (indirectly) linked to Complex I by the corresponding dehydrogenase-catalyzed reactions reducing NAD+ to NADH+H+ + H+. The most commonly applied N-junction substrate combinations are: PM, GM, PGM. The malate-anaplerotic pathway control state (M alone) is a special case related to malic enzyme (mtME). The glutamate-anaplerotic pathway control state (G alone) supports respiration through glutamate dehydrogenase (mtGDH). Oxidation of tetrahydrofolate is a NAD(P)H linked pathway with formation of formate. In mt-preparations, succinate dehydrogenase (SDH; CII) is largely substrate-limited in N-linked respiration, due to metabolite depletion into the incubation medium. The residual involvement of S-linked respiration in the N-pathway control state can be further suppressed by the CII-inhibitor malonic acid). In the N-pathway control state ET pathway level 4 is active. |
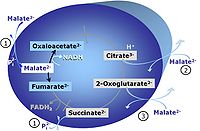
| NS-pathway control state | NS, CI&II | NS-pathway control is exerted in the NS-linked substrate state (flux in the NS-linked substrate state, NS; or Complex I&II, CI&II-linked substrate state). NS-OXPHOS capacity provides an estimate of physiologically relevant maximum mitochondrial respiratory capacity. NS is induced in mt-preparations by addition of NADH-generating substrates (N-pathway control state in combination with succinate (Succinate pathway; S). Whereas NS expresses substrate control in terms of substrate types (N and S), CI&II defines the same concept in terms of convergent electron transfer to the Q-junction (pathway control). NS is the abbreviation for the combination of NADH-linked substrates (N) and succinate (S). This physiological substrate combination is required for partial reconstitution of TCA cycle function and convergent electron-input into the Q-junction, to compensate for metabolite depletion into the incubation medium. NS in combination exerts an additive effect of convergent electron flow in most types of mitochondria. |
| NSGp | NSGp |
MitoPathway control state: NSGp
SUIT protocol: SUIT-038 This substrate combination supports convergent electron flow to the Q-junction. |
| OXPHOS capacity | P | |
| OctGM | OctGM | OctGM: Octanoylcarnitine & Glutamate & Malate.
MitoPathway control state: FN SUIT protocols: SUIT-015, SUIT-016, SUIT-017 |
| OctGMS | OctGMS | OctGMS: Octanoylcarnitine &Glutamate & Malate& Succinate.
MitoPathway control state: FNS SUIT protocols: SUIT-016, SUIT-017 |
| OctM pathway control state | OctM | OctM: Octanoylcarnitine & Malate.
MitoPathway control state: F SUIT protocols: SUIT-002, SUIT-015, SUIT-016, SUIT-017 Respiratory stimulation of the FAO-pathway, F, by fatty acid FA in the presence of malate M. Malate is a type N substrate (N), required for the F-pathway. In the presence of anaplerotic pathways (e.g., mitochondrial malic enzyme, mtME) the F-pathway capacity is overestimated, if there is an added contribution of NADH-linked respiration, F(N) (see SUIT-002). The FA concentration has to be optimized to saturate the FAO-pathway, without inhibiting or uncoupling respiration. Low concentration of malate, typically 0.1 mM, does not saturate the N-pathway; but saturates the F-pathway. High concentration of malate, typically 2 mM, saturates the N-pathway. |
| OctPGM pathway control state | OctPGM | OctPGM: Octanoylcarnitine & Pyruvate & Glutamate & Malate.
MitoPathway control state: FN SUIT protocols: SUIT-002
|
| OctPGMS pathway control state | OctPGMS | OctPGMS: Octanoylcarnitine & Pyruvate & Glutamate & Malate & Succinate.
MitoPathway control state: FNS SUIT protocol: SUIT-001, SUIT-002, SUIT-015 This substrate combination supports convergent electron flow to the Q-junction. |
| OctPGMSGp pathway control state | OctPGMSGp | OctPGMSGp: Octanoylcarnitine & Pyruvate & Glutamate & Malate & Succinate & Glycerophosphate.
MitoPathway control state: FNSGp SUIT protocol: SUIT-002 This substrate combination supports convergent electron flow to the Q-junction. |
| OctPM pathway control state | OctPM | OctPM: Octanoylcarnitine & Pyruvate & Malate.
MitoPathway control state: FN SUIT protocol: SUIT-002, SUIT-005 This substrate combination supports N-linked flux which is typically higher than FAO capacity (F/FN<0 in the OXPHOS state). In SUIT-RP1, PMOct is induced after PM(E), to evaluate any additive effect of adding Oct. In SUIT-RP2, FAO OXPHOS capacity is measured first, testing for the effect of increasing malate concentration (compare malate-anaplerotic pathway control state, M alone), and pyruvate is added to compare FAO as the background state with FN as the reference state. |
| OctPMS | OctPMS | OctPMS: Octanoylcarnitine & Pyruvate & Malate & Succinate.
MitoPathway control state: FNS SUIT protocol: SUIT-005 |
| PGM-pathway control state | PGM | PGM: Pyruvate & Glutamate & Malate.
MitoPathway control state: NADH electron transfer-pathway state Pyruvate (P) is oxidatively decarboxylated to acetyl-CoA and CO2, yielding NADH catalyzed by pyruvate dehydrogenase. Malate (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). Glutamate&malate is a substrate combination supporting an N-linked pathway control state, when glutamate is transported into the mt-matrix via the glutamate-aspartate carrier and reacts with oxaloacetate in the transaminase reaction to form aspartate and oxoglutarate. Glutamate as the sole substrate is transported by the electroneutral glutamate-/OH- exchanger, and is oxidized in the mitochondrial matrix by glutamate dehydrogenase to α-ketoglutarate ( 2-oxoglutarate), representing the glutamate-anaplerotic pathway control state. 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase, from oxaloacetate and glutamate by the transaminase, and from glutamate by the glutamate dehydrogenase. |
| PGMS-pathway control state | PGMS | PGMS: Pyruvate & Glutamate & Malate & Succinate.
MitoPathway control state: NS-pathway control state 2-oxoglutarate is produced through the citric acid cycle from citrate by isocitrate dehydrogenase, from oxaloacetate and glutamate by the transaminase, and from glutamate by the glutamate dehydrogenase. If the 2-oxoglutarate carrier does not outcompete these sources of 2-oxoglutarate, then the TCA cycle operates in full circle with external pyruvate&malate&glutamate&succinate |
| PGMSGp pathway control state | PGMSGp | PGMSGp: Pyruvate & Glutamate & Malate & Succinate & Glycerophosphate.
MitoPathway control state: NSGp SUIT protocol: SUIT-038 This substrate combination supports convergent electron flow to the Q-junction. |
| PM-pathway control state | PM | PM: Pyruvate & Malate.
MitoPathway control state: NADH Electron transfer-pathway state
|
| PMS-pathway control state | PMS | PMS: Pyruvate & Malate & Succinate.
MitoPathway control: CI&II Pyruvate (P) is oxidatively decarboxylated to acetyl-CoA and CO2, yielding NADH catalyzed by pyruvate dehydrogenase. Malate (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). This documents an additive effect of convergent CI&II electron flow to the Q-junction, with consistent results obtained with permeabilized muscle fibres and isolated mitochondria (Gnaiger 2009). |
| PalM | PalM | PalM: Palmitoylcarnitine & Malate.
MitoPathway control state: Fatty acid oxidation pathway control state SUIT protocols: SUIT-019 |
| PalOctM | PalOctM | PalOctM: Palmitoylcarnitine & Octanoylcarnitine & Malate.
MitoPathway control state: Fatty acid oxidation pathway control state SUIT protocols: SUIT-019 |
| PalOctPGM | PalOctPGM | PalOctPGM: Palmitoylcarnitine & Octanoylcarnitine & Pyruvate & Glutamate & Malate.
MitoPathway control state: FN SUIT protocols: SUIT-019 |
| PalOctPGMS | PalOctPGMS | PalOctPGMS: Palmitoylcarnitine & Octanoylcarnitine & Pyruvate & Glutamate & Malate & Succinate.
MitoPathway control state: FNS SUIT protocols: SUIT-019 |
| PalOctPM | PalOctPM | PalOctPM: Palmitoylcarnitine & Octanoylcarnitine & Pyruvate & Malate.
MitoPathway control state: FN SUIT protocols: SUIT-019 |
| PalPGMSGp pathway control state | PalPGMSGp | PalPGMSGp: Palmitoylcarnitine & Pyruvate & Glutamate & Malate & Succinate & Glycerophosphate.
MitoPathway control state: FNSGp SUIT protocol: SUIT-026 This substrate combination supports convergent electron flow to the Q-junction. |
| SGp-pathway control state | SGp | SGp: Succinate & Glycerophosphate.
MitoPathway control state: SGp; obtained with OctPGMSGp(Rot) SUIT protocol: SUIT-001 and ((SUIT-002 |
| Succinate pathway | S, SRot |
The Succinate pathway (S-pathway; S) is the electron transfer pathway that supports succinate-linked respiration (succinate-induced respiratory state; previously used nomenclature: CII-linked respiration; SRot; see Gnaiger 2009 Int J Biochem Cell Biol). The S-pathway describes the electron flux through Complex II (CII; see succinate dehydrogenase, SDH) from succinate and FAD to fumarate and CII-bound flavin adenine dinucleotide (FADH2) to the Q-junction. The S-pathway control state is usually induced in mt-preparations by addition of succinate&rotenone. In this case, only Complex III and Complex IV are involved in pumping protons from the matrix (positive phase, P-phase) to the negative phase (N-phase) with a P»/O2 of 3.5 (P»/O ratio = 1.75). |
- » SUIT concept
SUIT concept
| Term | Abbreviation | Description |
|---|---|---|
| Categories of SUIT protocols | SUIT-catg |
Categories of SUIT protocols group SUIT protocols according to all substrate types involved in a protocol (F, N, S, Gp), independent of the sequence of titrations of substrates and inhibitors which define the Electron-transfer-pathway states. The N-type substrates are listed in parentheses, independent of the sequence of titrations. ROX states may or may not be included in a SUIT protocol, which does not change its category. Similarly, the CIV assay may or may not be added at the end of a SUIT protocol, without effect on the category of a SUIT protocol.
|
| Cell respiration | Cell respiration channels metabolic fuels into the chemiosmotic coupling (bioenergetic) machinery of oxidative phosphorylation, being regulated by and regulating oxygen consumption (or consumption of an alternative final electron acceptor) and molecular redox states, ion gradients, mitochondrial (or microbial) membrane potential, the phosphorylation state of the ATP system, and heat dissipation in response to intrinsic and extrinsic energy demands. See also respirometry. In internal or cell respiration in contrast to fermentation, redox balance is maintained by external electron acceptors, transported into the cell from the environment. The chemical potential between electron donors and electron acceptors drives the electron transfer pathway, generating a chemiosmotic potential that in turn drives ATP synthesis. | |
| Coupling-control protocol | CCP | A coupling-control protocol CCP induces different coupling control states at a constant electron-transfer-pathway state. Residual oxygen consumption (Rox) is finally evaluated for Rox correction of flux. The CCP may be extended, when further respiratory states (e.g. cell viability test; CIV assay) are added to the coupling control module consisting of three coupling control states. The term phosphorylation control protocol, PCP, has been introduced synonymous for CCP. » MiPNet article |
| Coupling-control state | CCS | Coupling-control states are defined in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, homogenates) as LEAK respiration, OXPHOS, and ET states, with corresponding respiration rates (L, P, E) in any electron-transfer-pathway state which is competent for electron transfer. These coupling states are induced by titration of ADP and uncouplers, and application of specific inhibitors of the phosphorylation pathway. In living cells, the coupling-control states are LEAK respiration, ROUTINE, and ET states of respiration with corresponding rates L, R, E, using membrane-permeable inhibitors of the phosphorylation system (e.g. oligomycin) and uncouplers (e.g. CCCP). Coupling-control protocols induce these coupling-control states sequentially at a constant electron-transfer-pathway state. |
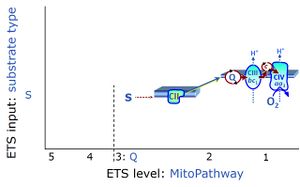
| Coupling/pathway control diagram | CPCD | Coupling/pathway control diagrams illustrate the respiratory states obtained step-by-step in substrate-uncoupler-inhibitor titrations in a SUIT protocol. Each step (to the next state) is defined by an initial state and a metabolic control variable, X. The respiratory states are shown by boxes. X is usually the titrated substance in a SUIT protocol. If X (ADP, uncouplers, or inhibitors of the phosphorylation system, e.g. oligomycin) exerts coupling control, then a transition is induced between two coupling-control states. If X (fuel substrates, e.g. pyruvate and succinate, or Electron transfer pathway inhibitors, e.g. rotenone) exerts pathway control, then a transition is induced between two Electron-transfer-pathway states. The type of metabolic control (X) is shown by arrows linking two respiratory states, with vertical arrows indicating coupling control, and horizontal arrows indicating pathway control. Marks define the section of an experimental trace in a given respiratory state (steady state). Events define the titration of X inducing a transition in the SUIT protocol. The specific sequence of coupling control and pathway control steps defines the SUIT protocol pattern. The coupling/pathway control diagrams define the categories of SUIT protocols (see Figure). |
| Cross-linked respiratory states | CLRS | Coordinated respiratory SUIT protocols are designed to include cross-linked respiratory states, which are common to these protocols. Different SUIT protocols address a variety of respiratory control steps which cannot be accomodated in a single protocol. Cross-linked respiratory states are included in each individual coordinated protocol, such that these states can be considered as replicate measurements, which also allow for harmonization of data obtained with these different protocols. |
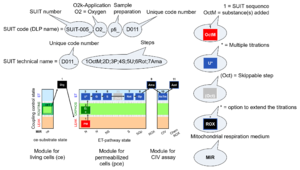
| D-number | D### | D number is the unique code given for each SUIT protocol. In the same SUIT protocol family (SUIT-###), there might be different protocols, specifically designed for different sample type (e.g., different mitochondrial preparations) or for different applications (e.g., O2, AmR, Fluo, MgG). Since the use of different kinds of sample or application may result in slightly different steps, each protocol receives a different D-number. |
| DatLab and SUIT protocols | This is a brief summary of steps to be taken for performing a high-resolution respirometry experiment with SUIT protocols using the OROBOROS Oroboros O2k and DatLab software. (1) Search for a specific SUIT protocol name (go to MitoPedia: SUIT). The list of MitoPedia SUIT protocols can be sorted by categories of SUIT protocols (sorting by SUIT protocol name), which is listed as the 'abbreviation' of the SUIT protocol name. (2) Copy the template for Mark names into your DatLab subdirectory: DatLab\APPDATA\MTEMPLAT. (3) Copy the DatLab-Analysis template for this SUIT protocol. (4) Follow the link to the corresponding publication or MiPNet communication, where the pdf file describing the SUIT protocol is available. (5) A DatLab demo file may be available providing an experimental example. After each sequential titration, a mark is set on the plot for flux or flow. After having set all marks, pull down the 'Mark names' menu, select the corresponding SUIT protocol for mark names, and rename all marks. The Mark names template also provides standard values of the titration volume preceding each mark. (6) Go to 'Mark statistics' [F2], copy to clipboard, and paste into the sample tab in the DatLab-Analysis template.
| |
| ET-pathway substrate types | n.a. | See Electron-transfer-pathway state |
| Electron-transfer-pathway state | ET-pathway state |
Electron-transfer-pathway states are obtained in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, tissue homogenate) by depletion of endogenous substrates and addition to the mitochondrial respiration medium of fuel substrates (CHNO) activating specific mitochondrial pathways, and possibly inhibitors of specific pathways. Mitochondrial electron-transfer-pathway states have to be defined complementary to mitochondrial coupling-control states. Coupling-control states require ET-pathway competent states, including oxygen supply. Categories of SUIT protocols are defined according to mitochondrial ET-pathway states. » MiPNet article |
| Ethanol | ethanol abs. |
Ethanol or ethyl alcohol, C2H6O or EtOH, is widely used in the laboratory, particularly as a solvent and cleaning agent. There are different grades of high purity ethanol. Up to a purity of 95.6 % ethanol can be separated from water by destillation. Higher concentrations than 95% require usage of additives that disrupt the azeotrope composition and allow further distillation. Ethanol is qualified as "absolute" if it contains no more than one percent water. Whenever 'ethanol abs.' is mentioned without further specification in published protocols, it refers to ≥ 99 % ethanol a.r. (analytical reagent grade).
|
| F-junction | The F-junction is a junction for convergent electron flow in the electron transfer pathway (ET-pathway) from fatty acids through fatty acyl CoA dehydrogenase (reduced form FADH2) to electron transferring flavoprotein (CETF), and further transfer through the Q-junction to Complex III (CIII). The concept of the F-junction and N-junction provides a basis for defining categories of SUIT protocols. Fatty acid oxidation, in the F-pathway control state, not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the N-junction substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCoA into the TCA cycle catalyzed by citrate synthase. | |
| Flux control efficiency | jZ-Y | Flux control efficiencies express the control of respiration by a metabolic control variable, X, as a fractional change of flux from YX to ZX, normalized for ZX. ZX is the reference state with high (stimulated or un-inhibited) flux; YX is the background state at low flux, upon which X acts.
Complementary to the concept of flux control ratios and analogous to elasticities of metabolic control analysis, the flux control efficiency of X upon background YX is expressed as the change of flux from YX to ZX normalized for the reference state ZX. » MiPNet article |
| Flux control ratio | FCR | Flux control ratios FCRs are ratios of oxygen flux in different respiratory control states, normalized for maximum flux in a common reference state, to obtain theoretical lower and upper limits of 0.0 and 1.0 (0 % and 100 %). For a given protocol or set of respiratory protocols, flux control ratios provide a fingerprint of coupling and substrate control independent of (1) mt-content in cells or tissues, (2) purification in preparations of isolated mitochondria, and (3) assay conditions for determination of tissue mass or mt-markers external to a respiratory protocol (CS, protein, stereology, etc.). FCR obtained from a single respirometric incubation with sequential titrations (sequential protocol; SUIT protocol) provide an internal normalization, expressing respiratory control independent of mitochondrial content and thus independent of a marker for mitochondrial amount. FCR obtained from separate (parallel) protocols depend on equal distribution of subsamples obtained from a homogenous mt-preparation or determination of a common mitochondrial marker. |
| Harmonized SUIT protocols | H-SUIT | Harmonized SUIT protocols (H-SUIT) are designed to include cross-linked respiratory states. When performing harmonized SUIT protocols in parallel, measurements of cross-linked respiratory states can be statistically evaluated as replicates across protocols. Additional information is obtained on respiratory coupling and substrate control by including respiratory states that are not common (not cross-linked) across the harmonized protocols. |
| Metabolic control variable | X | A metabolic control variable X causes the transition between a background state Y (background rate YX) and a reference state Z (reference rate ZX). X may be a stimulator or activator of flux, inducing the step change from background to reference steady state (Y to Z). Alternatively, X may be an inhibitor of flux, absent in the reference state but present in the background state (step change from Z to Y). |
| MitoFit protocols | MitoFit protocols are moderated by the MitoFit moderators (MitoFit team), either as protocols with direct reference to publications available to the scientific communicty, or protocols additionally described and made available in Bioblast with full information on authors (including contact details), author contributions, and editor (moderator) in charge. This aims at a comprehensive MitoFit data repository, which will require global input and cooperation. | |
| N-junction | The N-junction is a junction for convergent electron flow in the electron transfer pathway (ET-pathway) from type N substrates (further details »N-pathway control state) through the mt-NADH pool to Complex I (CI), and further transfer through the Q-junction to Complex III (CIII). Representative type N substrates are pyruvate (P), glutamate (G) and malate (M). The corresponding dehydrogenases (PDH, GDH, MDH) and some additional TCA cycle dehydrogenases (isocitrate dehydrogenase, oxoglutarate dehydrogenase generate NADH, the substrate of Complex I (CI). The concept of the N-junction and F-junction provides a basis for defining categories of SUIT protocols based on Electron-transfer-pathway states. | |
| NS e-input | NS, CI&II | NS e-input or the NS-pathway control state is electron input from a combination of substrates for the N-pathway control state and S-pathway control state through Complexes CI and CII simultaneously into the Q-junction. NS e-input corresponds to TCA cycle function in vivo, with convergent electron flow through the Electron transfer pathway. In mt-preparations, NS e-input requires addition not only of NADH- (N-) linked substrates (pyruvate&malate or glutamate&malate), but of succinate (S) simultaneously, since metabolite depletion in the absence of succinate prevents a significant stimulation of S-linked respiration. For more details, see: Additive effect of convergent electron flow. |
| OXPHOS capacity | P | |
| Phosphorylation pathway | DT | The phosphorylation pathway (phosphorylation system) is the functional unit utilizing the protonmotive force to phosphorylate ADP (D) to ATP (T), and may be defined more specifically as the P»-system. The P»-system consists of adenine nucleotide translocase, phosphate carrier, and ATP synthase. Mitochondrial adenylate kinase, mt-creatine kinase and mt-hexokinase constitute extended components of the P»-system, controlling local AMP and ADP concentrations and forming metabolic channels. Since substrate-level phosphorylation is involved in the TCA-cycle, the P»-system includes succinyl-CoA ligase (GDP to GTP or ADP to ATP). |
| Physiological pathway-control state | See Electron-transfer-pathway state. | |
| Preparation of SUIT chemicals | Preparation of SUIT chemicals describes the preparation of chemicals used in Substrate-Uncoupler-Inhibitor Ttitration (SUIT) protocols. | |
| Q-junction | The Q-junction is a junction for convergent electron flow in the Electron transfer pathway (ET-pathway) from type N substrates and mt-matrix dehydrogenases through Complex I (CI), from type F substrates and FA oxidation through electron-transferring flavoprotein Complex (CETF), from succinate (S) through Complex II (CII), from glycerophosphate (Gp) through glycerophosphate dehydrogenase Complex (CGpDH), from choline through choline dehydrogenase, from dihydro-orotate through dihydro-orotate dehydrogenase, and other enzyme Complexes into the Q-cycle (ubiquinol/ubiquinone), and further downstream to Complex III (CIII) and Complex IV (CIV). The concept of the Q-junction, with the N-junction and F-junction upstream, provides the rationale for defining Electron-transfer-pathway states and categories of SUIT protocols. | |
| Respiratory state | Respiratory states of mitochondrial preparations and living cells are defined in the current literature in many ways and with a diversity of terms. Mitochondrial respiratory states must be defined in terms of both, the coupling-control state and the electron-transfer-pathway state. | |
| SUIT | SUIT | SUIT is the abbreviation for Substrate-Uncoupler-Inhibitor Titration. SUIT protocols are used with mt-preparations to study respiratory control in a sequence of coupling and pathway control states induced by multiple titrations within a single experimental assay. These studies use biological samples economically to gain maximum information with a minimum amount of cells or tissue. |
| SUIT protocol library | SUITs | The Substrate-uncoupler-inhibitor titration (SUIT) protocol library contains a sequential list of SUIT protocols (D001, D002, ..) with links to the specific SUIT pages. Classes of SUIT protocols are explained with coupling and substrate control defined for mitochondrial preparations. |
| SUIT protocol names | SUITp-Names |
The SUIT protocol name starts with (i) the SUIT category which shows the Electron-transfer-pathway states (ET pathway types; e.g. N, S, NS, FNS, FNSGp), independent of the actual sequence of titrations. (ii) A further distinction is provided in the SUIT name by listing in parentheses the substrates applied in the N-pathway control states, again independent of the sequence of titrations, e.g. NS(GM), NS(PM), FNSGp(PGM). (iii) A sequentially selected number is added, e.g. SUIT_FNS(PM)01 (see Coupling/pathway control diagram). The systematic name of a SUIT protocol starts with the SUIT category, followed by an underline dash and the sequence of titration steps (mark names, #X, separated by a comma). The Marks define the section of a respiratory state in the SUIT protocol. The Mark name contains the sequential number and the metabolic control variable, X. The metabolic control variable is the name of the preceding SUIT event. The MitoPedia list of SUIT protocols can be sorted by the short name or the systematic name (hence by SUIT protocol category. The SUIT protocol pattern is best illustrated by a coupling/pathway control diagram. |
| SUIT protocol pattern | SUITp-Pattern | The SUIT protocol pattern describes the type of the sequence of coupling and substrate control steps in a SUIT protocol, which may be liner, orthogonal, or diametral. |
| SUIT reference protocol | SUIT RP | The substrate-uncoupler-inhibitor titration (SUIT) reference protocol, SUIT RP, provides a common baseline for comparison of mitochondrial respiratory control in a large variety of species, tissues and cell types, mt-preparations and laboratories, for establishing a database on comparative mitochondrial phyisology. The SUIT RP consists of two harmonized SUIT protocols (SUIT-001 - RP1 and SUIT-002 - RP2). These are coordinated such that they can be statistically evaluated as replicate measurements of cross-linked respiratory states, while additional information is obtained when the two protocols are conducted in parallel. Therefore, these harmonized SUIT protocols are complementary with their focus on specific respiratory coupling and pathway control aspects, extending previous strategies for respirometrc OXPHOS analysis.
|
| SUITbrowser | Use the SUITbrowser to find the substrate-uncoupler-inhibitor-titration (SUIT) protocol most suitable for addressing your research questions.
Open the SUITbrowser: http://suitbrowser.oroboros.at/
 How to find a DL-Protocol (DLP) How to find a DL-Protocol (DLP) | |
| Substrate control state | See Electron-transfer-pathway state | |
| Substrate-uncoupler-inhibitor titration | SUIT | Mitochondrial Substrate-uncoupler-inhibitor titration (SUIT) protocols are used with mitochondrial preparations to study respiratory control in a sequence of coupling and substrates states induced by multiple titrations within a single experimental assay. |
- See also: