Difference between revisions of "SUIT-020 Fluo mt D033"
| (45 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr=NS(PGM) | |abbr=NS(PGM) | ||
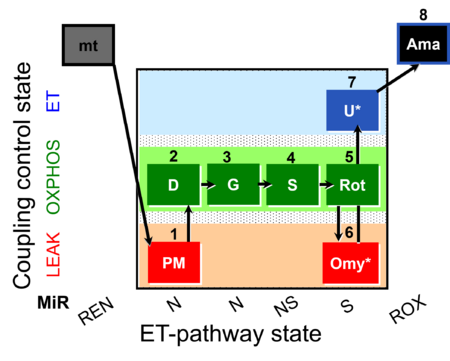
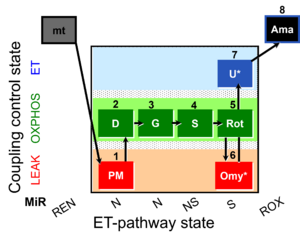
|description=[[File: | |description=[[File:1PM;2D;3G;4S;5Rot;6Omy;7U;8Ama.png|450px]] | ||
|info='''A''': | |info='''A''':short protocol for simultaneous determination of O<sub>2</sub> flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)- '''[[SUIT-020]]''' | ||
|application= | |application=Fluo | ||
|SUIT number=D033_1PM;2D;3G;4S;5Rot;6Omy;7U;8Ama | |SUIT number=D033_1PM;2D;3G;4S;5Rot;6Omy;7U;8Ama | ||
}} | }} | ||
::: '''[[Categories of SUIT protocols |SUIT-category]]:''' NS(PGM) | ::: '''[[Categories of SUIT protocols |SUIT-category]]:''' NS(PGM) | ||
::: '''[[SUIT protocol pattern]]:''' orthogonal 1PM;2D;3G;4S;5Rot;6Omy;7U | ::: '''[[SUIT protocol pattern]]:''' orthogonal 1PM;2D;3G;4S;5Rot;6Omy;7U- | ||
{{Template:SUIT text D033}} | |||
__TOC__ | __TOC__ | ||
Communicated by [[Komlodi T]] | Communicated by [[Huete-Ortega M]], [[Antunes D]], [[Komlodi T]] and [[Gnaiger E]] (last update 2019-07-25) | ||
== | |||
: | == Representative traces == | ||
:::* [[SUIT- | |||
[[File:SUIT-020 Fluo mt D033 O2 DL-File2.png|600px]] | |||
[[File:SUIT-020 Fluo mt D033 DL-File2.png|600px]] | |||
{{Template:SUIT-020 Fluo mt D033}} | |||
== Strengths and limitations == | |||
:::* Before performing this protocol, a calibration with the fluoresence dye needs to be done. More information on our USB: Instrumental Protocols/Fluo calibration. | |||
:::* It is recommended to run a chemical background without any sample to test the effect of the chemicals on the fluorescence signal. More information on our USB. | |||
:::* NS-OXPHOS capacity provides a physiologically relevant estimate of maximum mitochondrial respiratory capacity. | |||
:::* Comparison of GM- with PM-capacity yields important information on N-pathway respiratory control upstream of CI. | |||
:::* A succinate concentration of >10 mM may be required for saturating SE capacity. | |||
:::* [[Nigericin]] as a H<sup>+</sup>/K<sup>+</sup> antiporter can be used to dissipate transmembrane pH gradient, which results in increased [[Mitochondrial membrane potential|mt-membrane potential]] in the LEAK state. | |||
:::+ You obtain information in a single protocol about the [[NS-pathway control state| NS-]], [[Succinate pathway control state| S]] and [[NADH Electron transfer-pathway state| N-]] pathway control state. | |||
:::- Many fluorescence dyes (such as Safranin, TMRM etc) can inhibit components of the ET system, most commonly affecting NADH-linked respiration. Therefore, a control run of the protocol should be done in the absence of the fluorescence dye. The following protocol can be used: [[SUIT-020 O2 mt D032]]. | |||
:::- To test the effect of the fluorescence dye on the respiration, in [[SUIT-020 O2 mt D032]] and [[SUIT-021 O2 mt D035]] the dye can be added after cytochrome c. | |||
:::- CIV activity cannot be determined and Cytochrome c test cannot be performed together with the fluorescence dyes. | |||
:::- [[Oligomycin| Oligomycin]] concentration has to be determined. Higher concentrations of Omy may inhibit the ET state. | |||
:::- Careful washing is required after the experiment to avoid carry-over of the inhibitors and uncoupler. | |||
== Compare SUIT protocols == | |||
:::* [[SUIT-004]];[[SUIT-004 O2 pfi D010]]: provides information about the NS(PM) pathway in the ET state without the contribution of G. | |||
:::* [[SUIT-008]];[[SUIT-008 O2 pfi D014]] and [[SUIT-008 O2 ce-pce D025]]: provides information about the NS(PGM) pathway in the OXPHOS and ET-states, but without the addition of Omy. | |||
:::* [[SUIT-011]]: provides information about the NS(GM) pathway in the OXPHOS and ET-states, but without the addition of Omy. | |||
:::* [[SUIT-012]]: provides information about the N(PGM) pathway in the OXPHOS and ET-states without the contribution of S-pathway and without the addition of Omy. | |||
:::* [[SUIT-014]]: a protocol similar to [[SUIT-021 O2 mt D035]], but with the addition of P in the OXPHOS state before S. Omy is not added in the protocol. | |||
== Chemicals and syringes == | |||
{{Template:Chemicals Saf}} | |||
or | |||
{{Template:Chemicals TMRM}} | |||
or | |||
{{Template:Chemicals Rh123}} | |||
{{Template:Chemicals SUIT-020}} | |||
::: Suggested stock concentrations are shown in the specific DL-Protocol. | |||
== References == | == References == | ||
| Line 33: | Line 67: | ||
{{#ask:[[Category:Abstracts]] [[Additional label::SUIT-020 Fluo mt D033]] | {{#ask:[[Category:Abstracts]] [[Additional label::SUIT-020 Fluo mt D033]] | ||
| ?Was submitted in year=Year | | ?Was submitted in year=Year | ||
| ?Has title=Reference | | ?Has title=Reference | ||
| Line 45: | Line 79: | ||
}} | }} | ||
{{MitoPedia concepts | {{MitoPedia concepts | ||
Latest revision as of 08:41, 30 September 2020
Description
Abbreviation: NS(PGM)
Reference: A:short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)- SUIT-020
SUIT number: D033_1PM;2D;3G;4S;5Rot;6Omy;7U;8Ama
O2k-Application: Fluo
- SUIT-category: NS(PGM)
- SUIT protocol pattern: orthogonal 1PM;2D;3G;4S;5Rot;6Omy;7U-
SUIT-020 Fluo mt D033 is a protocol to assess the additivity between N-pathway and S- pathway in the Q-junction as well as investigate the N- and NS-pathway control state in mitochondrial preparations. It can serve as a diagnostic tool for the activity of glutamate dehydrogenase and its linked pathway. Oligomycin (Omy) is used to induce a LEAK state of respiration via the inhibition of the ATP synthase. Since higher concentrations of Omy can decrease the ET state induced upon addition of uncoupler, the required concentration of Omy has to be assessed by the Omy titration test. Addition of PM (Pyruvate & Malate) to the mitochondria leads to the hyperpolarisation of the mt-membrane, while ADP (D) decreases the mt-membrane potential. Glutamate (G) and Succinate (S) are able to further increase the mt-membrane potential. Rotenone, which is an inhibitor of Complex I, depolarises the mt-membrane. Addition of Omy results in hyperpolarisation owing to the fact that the inhibition of the ATP synthase leads to accumulation of protons in the intermembrane space. Uncoupler depolarises the mt-membrane in a concentration-dependent manner, therefore leading to dissipation of the mt-membrane potential. Complex III inhibitor Antimycin A (Ama) blocks the respiration and dissipates the mt-membrane potential. For mt-membrane potential analysis and calculation, visit this page: Mitochondrial membrane potential.
Communicated by Huete-Ortega M, Antunes D, Komlodi T and Gnaiger E (last update 2019-07-25)
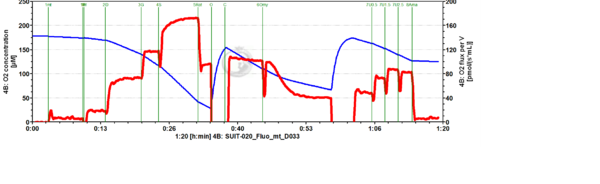
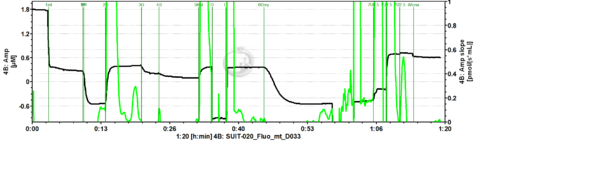
Representative traces
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1PM | PML(n) | N | CI | 1PM
|
| 2D | PMP | N | CI | 1PM;2D
|
| 3G | PGMP | N | CI | 1PM;2D;3G
|
| 4S | PGMSP | NS | CI&II | 1PM;2D;3G;4S
|
| 5Rot | SP | S | CII | 1PM;2D;3G;4S;5Rot
|
| 6Omy | SL(Omy) | 1PM;2D;3G;4S;5Rot;6Omy
| ||
| 7U* | SE | S | CII | 1PM;2D;3G;4S;5Rot;6Omy;7S
|
| 8Ama | ROX | 1PM;2D;3G;4S;5Rot;6Omy;7S;8Ama
|
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Strengths and limitations
- Before performing this protocol, a calibration with the fluoresence dye needs to be done. More information on our USB: Instrumental Protocols/Fluo calibration.
- It is recommended to run a chemical background without any sample to test the effect of the chemicals on the fluorescence signal. More information on our USB.
- NS-OXPHOS capacity provides a physiologically relevant estimate of maximum mitochondrial respiratory capacity.
- Comparison of GM- with PM-capacity yields important information on N-pathway respiratory control upstream of CI.
- A succinate concentration of >10 mM may be required for saturating SE capacity.
- Nigericin as a H+/K+ antiporter can be used to dissipate transmembrane pH gradient, which results in increased mt-membrane potential in the LEAK state.
- + You obtain information in a single protocol about the NS-, S and N- pathway control state.
- - Many fluorescence dyes (such as Safranin, TMRM etc) can inhibit components of the ET system, most commonly affecting NADH-linked respiration. Therefore, a control run of the protocol should be done in the absence of the fluorescence dye. The following protocol can be used: SUIT-020 O2 mt D032.
- - To test the effect of the fluorescence dye on the respiration, in SUIT-020 O2 mt D032 and SUIT-021 O2 mt D035 the dye can be added after cytochrome c.
- - CIV activity cannot be determined and Cytochrome c test cannot be performed together with the fluorescence dyes.
- - Oligomycin concentration has to be determined. Higher concentrations of Omy may inhibit the ET state.
- - Careful washing is required after the experiment to avoid carry-over of the inhibitors and uncoupler.
Compare SUIT protocols
- SUIT-004;SUIT-004 O2 pfi D010: provides information about the NS(PM) pathway in the ET state without the contribution of G.
- SUIT-008;SUIT-008 O2 pfi D014 and SUIT-008 O2 ce-pce D025: provides information about the NS(PGM) pathway in the OXPHOS and ET-states, but without the addition of Omy.
- SUIT-011: provides information about the NS(GM) pathway in the OXPHOS and ET-states, but without the addition of Omy.
- SUIT-012: provides information about the N(PGM) pathway in the OXPHOS and ET-states without the contribution of S-pathway and without the addition of Omy.
- SUIT-014: a protocol similar to SUIT-021 O2 mt D035, but with the addition of P in the OXPHOS state before S. Omy is not added in the protocol.
Chemicals and syringes
| Step | Chemical(s) and link(s) | Comments |
|---|---|---|
| Saf | Safranin (Saf) |
or
| Step | Chemical(s) and link(s) | Comments |
|---|---|---|
| TMRM | Tetramethylrhodamine methyl ester (TMRM) |
or
| Step | Chemical(s) and link(s) | Comments |
|---|---|---|
| Rh123 | Rhodamine 123 |
| Step | Chemical(s) and link(s) | Comments |
|---|---|---|
| 1PM | Pyruvate (P) and Malate (M) | |
| 2D | ADP (D) | |
| 3G | Glutamate (G) | |
| 4S | Succinate (S) | |
| 5Rot | Rotenone (Rot) | |
| 6Omy | Oligomycin (Omy) | |
| 7U | Carbonyl cyanide m-chlorophenyl hydrazone, CCCP (U) | Can be substituted for other uncoupler |
| 8Ama | Antimycin A (Ama) |
- Suggested stock concentrations are shown in the specific DL-Protocol.
References
| Year | Reference | Organism | Tissue;cell | |
|---|---|---|---|---|
| MiPNet20.13 Safranin mt-membranepotential | 2019-06-24 | O2k-FluoRespirometry: HRR and simultaneous determination of mt-membrane potential with safranin or TMRM. | Mouse | Nervous system |
| Krumschnabel 2014 Methods Enzymol | 2014 | Krumschnabel G, Eigentler A, Fasching M, Gnaiger E (2014) Use of safranin for the assessment of mitochondrial membrane potential by high-resolution respirometry and fluorometry. Methods Enzymol 542:163-81. https://doi.org/10.1016/B978-0-12-416618-9.00009-1 | Mouse | Nervous system |
MitoPedia concepts:
SUIT protocol,
SUIT A,
Find
MitoPedia methods:
Fluorometry