Difference between revisions of "SUIT-013 AmR ce D023"
From Bioblast
Timon Alba (talk | contribs) |
|||
| (7 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr=ce | |abbr=O<sub>2 dependence of H<sub>2</sub>O<sub>2</sub> production ce | ||
|description=[[File:SUIT013 AmR ce D023.png| | |description=[[File:SUIT013 AmR ce D023.png|400px]] | ||
|info='''A''' '''[[SUIT-013]]''' | |info='''A''' '''[[SUIT-013]]''' | ||
|application=AmR | |application=AmR | ||
| Line 7: | Line 7: | ||
}} | }} | ||
::: '''[[MitoPedia: SUIT]]''' | ::: '''[[MitoPedia: SUIT]]''' | ||
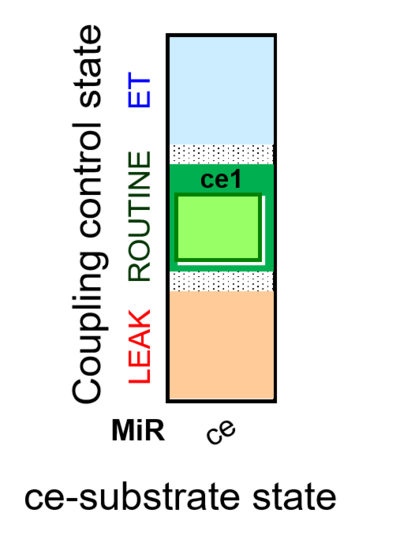
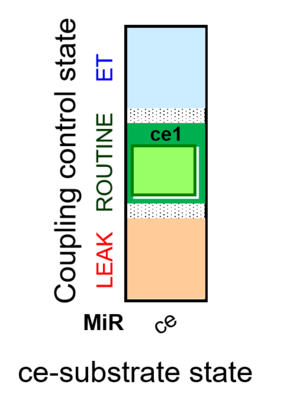
::: '''[[SUIT protocol pattern]]:''' ce1 | |||
::: '''[[SUIT protocol pattern]]:''' | |||
This protocol has been designed to study the oxygen dependence of H<sub>2</sub>O<sub>2</sub> production in | This protocol has been designed to study the oxygen dependence of H<sub>2</sub>O<sub>2</sub> production in living cells. SUIT-013 AmR ce D023 is suitable for demonstration of the oxygen dependence in baker´s yeast. For further details see: [[Komlodi 2021 MitoFit AmR-O2]]. The respiration medium has to be chosen carefully, since the [[Amplex UltraRed]] assay leads to artefact formation with the components of the buffer. | ||
__TOC__ | __TOC__ | ||
Communicated by [[Komlodi T]] | Communicated by [[Komlodi T]] (last update 2024-03-07) | ||
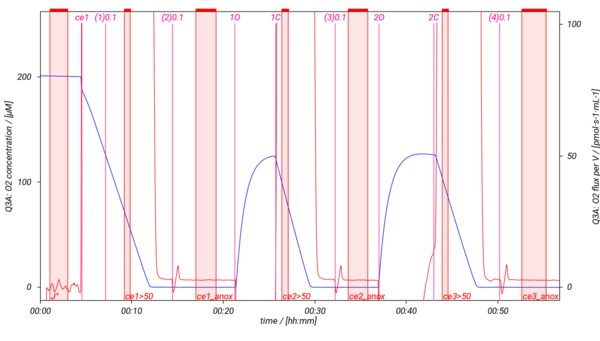
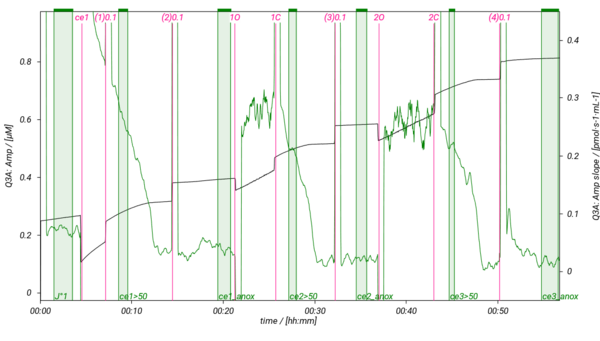
== Representative traces == | |||
[[File:SUIT-013 O2 D023.png|600px]] | |||
[[File:SUIT-013 AmR D023.png|600px]] | |||
{{Template:SUIT-013}} | {{Template:SUIT-013}} | ||
Latest revision as of 11:31, 7 March 2024
Description
Abbreviation: O2 dependence of H2O2 production ce
Reference: A SUIT-013
SUIT number: D023_ce1
O2k-Application: AmR
This protocol has been designed to study the oxygen dependence of H2O2 production in living cells. SUIT-013 AmR ce D023 is suitable for demonstration of the oxygen dependence in baker´s yeast. For further details see: Komlodi 2021 MitoFit AmR-O2. The respiration medium has to be chosen carefully, since the Amplex UltraRed assay leads to artefact formation with the components of the buffer.
Communicated by Komlodi T (last update 2024-03-07)
Representative traces
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 0DTPA |
| |||
| 0SOD |
| |||
| 0HRP |
| |||
| 0AmR |
|
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| ce1 | ROUTINE | ce1
|
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Strengths and limitations
- To study the inhibitory effect of the AmR assay on the respiration using living cells, the following protocols are recommended: SUIT 003 AmR ce D058 with AmR and SUIT 003 AmR ce D059 with carrier titration as a control. These two protocols have to be done in parallel.
- + Reasonable duration of the experiment.
- + Quick demonstration of the oxygen dependence of H2O2 production.
- + No externally added substrates are required with yeast.
Compare SUIT protocols
References
| Year | Reference | Organism | Tissue;cell | |
|---|---|---|---|---|
| MiPNet24.10 H2O2 flux analysis | 2021-10-22 | Hydrogen peroxide flux analysis using Amplex UltraRed assay in MiR05-Kit with DatLab 7.4 | ||
| Komlodi 2021 BEC AmR-O2 | 2021 | Komlódi T, Sobotka O, Gnaiger E (2021) Facts and artefacts on the oxygen dependence of hydrogen peroxide production using Amplex UltraRed. Bioenerg Commun 2021.4. https://doi.org/10.26124/bec:2021-0004 | Saccharomyces cerevisiae | Other cell lines |
| MiPNet20.14 AmplexRed H2O2-production | 2019-06-24 | O2k-FluoRespirometry: HRR and simultaneous determination of H2O2 production with Amplex UltraRed. | Mouse | Heart |
| Komlodi 2018 Methods Mol Biol | 2018 | Komlodi T, Sobotka O, Krumschnabel G, Bezuidenhout N, Hiller E, Doerrier C, Gnaiger E (2018) Comparison of mitochondrial incubation media for measurement of respiration and hydrogen peroxide production. Methods Mol Biol 1782:137-55. | Human Mouse | Skeletal muscle HEK |
| Makrecka-Kuka 2015 Biomolecules | 2015 | Makrecka-Kuka M, Krumschnabel G, Gnaiger E (2015) High-resolution respirometry for simultaneous measurement of oxygen and hydrogen peroxide fluxes in permeabilized cells, tissue homogenate and isolated mitochondria. https://doi.org/10.3390/biom5031319 | Human Mouse | Heart Nervous system HEK |
| Krumschnabel 2015 Methods Mol Biol | 2015 | Krumschnabel G, Fontana-Ayoub M, Sumbalova Z, Heidler J, Gauper K, Fasching M, Gnaiger E (2015) Simultaneous high-resolution measurement of mitochondrial respiration and hydrogen peroxide production. Methods Mol Biol 1264:245-61. | Mouse | Nervous system |
MitoPedia concepts: SUIT protocol, SUIT A, Find
MitoPedia methods:
Respirometry,
Fluorometry