Difference between revisions of "SUIT-020"
From Bioblast
| Line 2: | Line 2: | ||
|description=[[File:SUIT-020 O2 mt D032.png|400px]] [[File:SUIT-020 O2 mt D035.png|400px]] | |description=[[File:SUIT-020 O2 mt D032.png|400px]] [[File:SUIT-020 O2 mt D035.png|400px]] | ||
|info='''A:'''[[File:PDF.jpg|100px|link=http://wiki.oroboros.at/images/6/64/SUIT-020.pdf|Bioblast pdf]] »[http://wiki.oroboros.at/index.php/File:SUIT-020.pdf Versions] | |info='''A:'''[[File:PDF.jpg|100px|link=http://wiki.oroboros.at/images/6/64/SUIT-020.pdf|Bioblast pdf]] »[http://wiki.oroboros.at/index.php/File:SUIT-020.pdf Versions] | ||
{{MitoPedia O2k and high-resolution respirometry}} | {{MitoPedia O2k and high-resolution respirometry}} | ||
{{MitoPedia topics}} | {{MitoPedia topics}} | ||
::: '''[[Categories of SUIT protocols |SUIT-category]]:''' | ::: '''[[Categories of SUIT protocols |SUIT-category]]:''' NS (GM) and NS(PGM) | ||
::: '''[[SUIT protocol pattern]]:''' | ::: '''[[SUIT protocol pattern]]:''' orthogonal 1PM;2D;3G;4S;5Rot;6Omy;7U;- and 1GM;2D;3S;4Rot;5Omy;6U;- | ||
__TOC__ | __TOC__ | ||
| Line 44: | Line 37: | ||
::: | ::: | ||
== Compare SUIT protocols == | |||
:::* [[SUIT-001]];[[SUIT-001 O2 mt D001]]for isolated mitochondria and tissue homogenate: a complex protocol to get information not only about the NS(PGM) in the OXPHOS state, but also in ET state and about the FGp-pathways additionally. | |||
:::* [[SUIT-004]];[[SUIT-004 O2 pfi D010]]: it provides an information about the NS(PM) pathway in the ET state withou the contribution of G. | |||
:::* [[SUIT-008]];[[SUIT-008 O2 pfi D014]] and [[SUIT-008 O2 pce D025]]: it provides information about the NS(PGM) pathway in the OXPHOS and ET-states, but without the addtion of Omy. | |||
:::* [[SUIT-011]]:it provides information about the NS(GM) pathway in the OXPHOS and ET-states, but without the addtion of Omy. | |||
:::* [[SUIT-012]]:it provides information about the N(PGM) pathway in the OXPHOS and ET-states wihtout the contribution of S-pathway and without the addtion of Omy. | |||
:::* [[SUIT-014]]: it is similar protocol to [[SUIT-020 O2 mt D035]], but with the addition of P in the OXPHOS state before S. Omy is not added in the protocol. | |||
= | }} | ||
{{MitoPedia concepts | |||
|mitopedia concept=MiP concept, SUIT protocol, SUIT A, Recommended | |||
}} | |||
{{MitoPedia methods | |||
|mitopedia method=Respirometry, Fluorometry | |||
}} | |||
Revision as of 13:02, 28 January 2019
Description
Reference: A:![]() »Versions
»Versions
- SUIT-category: NS (GM) and NS(PGM)
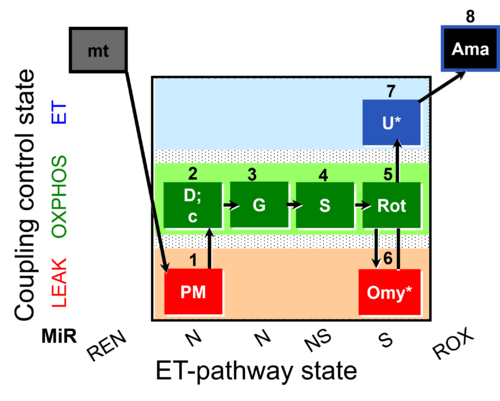
- SUIT protocol pattern: orthogonal 1PM;2D;3G;4S;5Rot;6Omy;7U;- and 1GM;2D;3S;4Rot;5Omy;6U;-
DLP applications
- SUIT-020 Fluo mt D033 simultaneous determination of O2 flux and mt-membrane potential using any fluorescence dye (Safranin, TMRM etc) on isolated mitochondria and tissue homogenate.
- SUIT-020 Fluo mt D036 simultaneous determination of O2 flux and mt-membrane potential using any fluorescence dye (Safranin, TMRM etc) on isolated mitochondria and tissue homogenate.
- SUIT-020 O2 mt D032 determination of O2 flux on isolated mitochondria and tissue homogenate as a control for SUIT-020 Safr mt D033.
- SUIT-020 O2 mt D035 determination of O2 flux on isolated mitochondria and tissue homogenate as a control for SUIT-020 Safr mt D036.
References
| Year | Reference | Organism | Tissue;cell | |
|---|---|---|---|---|
| MiPNet20.13 Safranin mt-membranepotential | 2019-06-24 | O2k-FluoRespirometry: HRR and simultaneous determination of mt-membrane potential with safranin or TMRM. | Mouse | Nervous system |
| Krumschnabel 2014 Methods Enzymol | 2014 | Krumschnabel G, Eigentler A, Fasching M, Gnaiger E (2014) Use of safranin for the assessment of mitochondrial membrane potential by high-resolution respirometry and fluorometry. Methods Enzymol 542:163-81. https://doi.org/10.1016/B978-0-12-416618-9.00009-1 | Mouse | Nervous system |
'"`UNIQ--h-1--QINU`"' Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1PM | PML(n) | N | CI | 1PM
|
| 2D | PMP | N | CI | 1PM;2D
|
| 2c | PMcP | N | CI | 1PM;2D;2c
|
| 3G | PGMP | N | CI | 1PM;2D;2c;3G
|
| 4S | PGMSP | NS | CI&II | 1PM;2D;2c;3G;4S
|
| 5Rot | SP | S | CII | 1PM;2D;2c;3G;4S;5Rot
|
| 6Omy | SL(Omy) | 1PM;2D;2c;3G;4S;5Rot;6Omy
| ||
| 7U* | SE | S | CII | 1PM;2D;2c;3G;4S;5Rot;6Omy;7S
|
| 8Ama | ROX | 1PM;2D;2c;3G;4S;5Rot;6Omy;7S;8Ama
|
| Step | Respiratory state | Pathway control | ET-Complex | Comment |
|---|---|---|---|---|
| ## AsTm | AsTmE | CIV | CIV | |
| ## Azd | CHB |
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Strengths and limitations
- + Information in one protocol about the NS, S and N-pathway control state.
- - The fluorescence dye (such as Safranin, TMRM etc) can inhibit most porbably the NADH-linked respiration, therefore, the protocol has to be run without using the fluorescence dye. The following protocols can be used: SUIT-020 O2 mt D032, SUIT-020 O2 mt D035.
Compare SUIT protocols
- SUIT-001;SUIT-001 O2 mt D001for isolated mitochondria and tissue homogenate: a complex protocol to get information not only about the NS(PGM) in the OXPHOS state, but also in ET state and about the FGp-pathways additionally.
- SUIT-004;SUIT-004 O2 pfi D010: it provides an information about the NS(PM) pathway in the ET state withou the contribution of G.
- SUIT-008;SUIT-008 O2 pfi D014 and SUIT-008 O2 pce D025: it provides information about the NS(PGM) pathway in the OXPHOS and ET-states, but without the addtion of Omy.
- SUIT-011:it provides information about the NS(GM) pathway in the OXPHOS and ET-states, but without the addtion of Omy.
- SUIT-012:it provides information about the N(PGM) pathway in the OXPHOS and ET-states wihtout the contribution of S-pathway and without the addtion of Omy.
- SUIT-014: it is similar protocol to SUIT-020 O2 mt D035, but with the addition of P in the OXPHOS state before S. Omy is not added in the protocol.
MitoPedia concepts:
MiP concept,
SUIT protocol,
SUIT A,
Recommended
MitoPedia methods:
Respirometry,
Fluorometry