The MitoPedia terminology is developed continuously in the spirit of Gentle Science.
| Term | Abbreviation | Description |
|---|---|---|
| Abscissa | x | The abscissa is the horizontal axis x of a rectangular two-dimensional graph with the ordinate y as the vertical axis. Values X are placed horizontally from the origin. See Abscissal X/Y regression. |
| Acclimation | Acclimation is an immediate time scale adaption expressing phenotypic plasticity in response to changes of a single variable under controlled laboratory conditions. | |
| Acclimatization | Acclimatization is an immediate time scale adaption expressing phenotypic plasticity in response to changes of habitat conditions and life style where several variables may change simultaneously. | |
| Adaptation | Adaptation is an evolutionary time scale expression of phenotypic plasticity in response to selective pressures prevailing under various habitat conditions. | |
| Advancement | dtrξ [MU] | In an isomorphic analysis, any form of flow is the advancement of a process per unit of time, expressed in a specific motive unit [MU∙s-1], e.g., ampere for electric flow or current, Iel = delξ/dt [A≡C∙s-1], watt for thermal or heat flow, Ith = dthξ/dt [W≡J∙s-1], and for chemical flow of reaction, Ir = drξ/dt, the unit is [mol∙s-1] (extent of reaction per time). The corresponding motive forces are the partial exergy (Gibbs energy) changes per advancement [J∙MU-1], expressed in volt for electric force, ΔelF = ∂G/∂elξ [V≡J∙C-1], dimensionless for thermal force, ΔthF = ∂G/∂thξ [J∙J-1], and for chemical force, ΔrF = ∂G/∂rξ, the unit is [J∙mol-1], which deserves a specific acronym [Jol] comparable to volt [V]. For chemical processes of reaction (spontaneous from high-potential substrates to low-potential products) and compartmental diffusion (spontaneous from a high-potential compartment to a low-potential compartment), the advancement is the amount of motive substance that has undergone a compartmental transformation [mol]. The concept was originally introduced by De Donder [1]. Central to the concept of advancement is the stoichiometric number, νi, associated with each motive component i (transformant [2]).
In a chemical reaction r the motive entity is the stoichiometric amount of reactant, drni, with stoichiometric number νi. The advancement of the chemical reaction, drξ [mol], is defined as, drξ = drni·νi-1 The flow of the chemical reaction, Ir [mol·s-1], is advancement per time, Ir = drξ·dt-1 This concept of advancement is extended to compartmental diffusion and the advancement of charged particles [3], and to any discontinuous transformation in compartmental systems [2], |
| Aerobic | ox | The aerobic state of metabolism is defined by the presence of oxygen (air) and therefore the potential for oxidative reactions (ox) to proceed, particularly in oxidative phosphorylation (OXPHOS). Aerobic metabolism (with involvement of oxygen) is contrasted with anaerobic metabolism (without involvement of oxygen): Whereas anaerobic metabolism may proceed in the absence or presence of oxygen (anoxic or oxic conditions), aerobic metabolism is restricted to oxic conditions. Below the critical oxygen pressure, aerobic ATP production decreases. |
| Ambiguity crisis |
The ambiguity crisis is a contemporary crisis comparable to the credibility or reproducibility crisis in the biomedical sciences. The term 'crisis' is rooted etymologically in the Greek word krinein: meaning to 'separate, decide, judge'. In this sense, science and communication in general are a continuous crisis at the edge of separating clarity or certainty from confusing double meaning, or obscure 'alchemical' gibberish, or even fake-news. Reproducibility relates to the condition of repeating and confirming calculations or experiments presented in a published resource. While ambiguity is linked to relevant issues of reproducibility, it extends to the communications space of terminological and graphical representations of concepts. Type 1 ambiguities are the inevitable consequence of conceptual evolution, in the process of which ambiguities are replaced by experimentally and theoretically supported paradigm shifts to clear-cut theorems. In contrast, type 2 ambiguities are traced in publications that reflect merely a disregard and ignorance of established concepts without an attempt to justify the inherent deviations from high-quality science. There are many shades of grey between these types of ambiguity. | |
| Amount of substance | n [mol] | The amount of substance n is a base physical quantity, and the corresponding SI unit is the mole [mol]. Amount of substance (sometimes abbreviated as 'amount' or 'chemical amount') is proportional to the number NX of specified elementary entities X, and the universal proportionality constant is the reciprocal value of the Avogadro constant (SI),
nX = NX·NA-1 nX contained in a system can change due to internal and external transformations, dnX = dinX + denX In the absence of nuclear reactions, the amount of any atom is conserved, e.g., for carbon dinC = 0. This is different for chemical substances or ionic species which are produced or consumed during the advancement of a reaction r, A change in the amount of Xi, dni, in an open system is due to both the internal formation in chemical transformations, drni, and the external transfer, deni, across the system boundaries. dni is positive if Xi is formed as a product of the reaction within the system. deni is negative if Xi flows out of the system and appears as a product in the surroundings (Cohen 2008 IUPAC Green Book). |
| Anaerobic | Anaerobic metabolism takes place without the use of molecular oxygen, in contrast to aerobic metabolism. The capacity for energy assimilation and growth under anoxic conditions is the ultimate criterion for facultative anaerobiosis. Anaerobic metabolism may proceed not only under anoxic conditions or states, but also under hyperoxic and normoxic conditions (aerobic glycolysis), and under hypoxic and microxic conditions below the limiting oxygen pressure. | |
| Anaplerosis | Anaplerosis is the process of formation of intermediates of the tricarboxylic acid cycle. Malic enzyme (mtME), phosphoenolpyruvate carboxykinase (PEPCK), propionyl-CoA carboxylase, pyruvate carboxylase and proline dehydrogenase play important roles in anaplerosis. | |
| Anoxia | anox | Ideally the terms anoxia and anoxic (anox, without oxygen) should be restricted to conditions where molecular oxygen is strictly absent. Practically, effective anoxia is obtained when a further decrease of experimental oxygen levels does not elicit any physiological or biochemical response. The practical definition, therefore, depends on (i) the techiques applied for oxygen removal and minimizing oxygen diffusion into the experimental system, (ii) the sensitivity and limit of detection of analytical methods of measuring oxygen (O2 concentration in the nM range), and (iii) the types of diagnostic tests applied to evaluate effects of trace amounts of oxygen on physiological and biochemical processes. The difficulties involved in defining an absolute limit between anoxic and microxic conditions are best illustrated by a logarithmic scale of oxygen pressure or oxygen concentration. In the anoxic state (State 5), any aerobic type of metabolism cannot take place, whereas anaerobic metabolism may proceed under oxic or anoxic conditions. |
| Artemisinin | Artemisinin and various derivatives are potent anti-malaria drugs which have additionally anti-tumorigenic effects, particularly when targeted at mitochondria. The anti-malaria effect is associated with artemisinin's action on heme. Mitochondria are involved in the synthesis of heme, and may play additional roles in the anti-tumorigenic effect of artemisinin. | |
| Aspirin | Aspirin is a widely applied drug that requires dosage adjusted to individual body mass. It is a non-selective COX inhibitor and exerts an effect on long-chain fatty acid transport into mitochondria. | |
| BME cutoff points | BME cutoff | Obesity is defined as a disease associated with an excess of body fat with respect to a healthy reference condition. Cutoff points for body mass excess, BME cutoff points, define the critical values for underweight (-0.1 and -0.2), overweight (0.2), and various degrees of obesity (0.4, 0.6, 0.8, and above). BME cutoffs are calibrated by crossover-points of BME with established BMI cutoffs. |
| Basal respiration | BMR | Basal respiration or basal metabolic rate (BMR) is the minimal rate of metabolism required to support basic body functions, essential for maintenance only. BMR (in humans) is measured at rest 12 to 14 hours after eating in a physically and mentally relaxed state at thermally neutral room temperature. Maintenance energy requirements include mainly the metabolic costs of protein turnover and ion homeostasis. In many aerobic organisms, and particularly well studied in mammals, BMR is fully aerobic, i.e. direct calorimetry (measurement of heat dissipation) and indirect calorimetry (measurement of oxygen consumption multiplied by the oxycaloric equivalent) agree within errors of measurement (Blaxter KL 1962. The energy metabolism of ruminants. Hutchinson, London: 332 pp [1]). In many cultured mammalian cells, aerobic glycolysis contributes to total ATP turnover (Gnaiger and Kemp 1990 [2]), and under these conditions, 'respiration' is not equivalent to 'metabolic rate'. Basal respiration in humans and skeletal muscle mitochondrial function (oxygen kinetics) are correlated (Larsen et al 2011 [3]). » MiPNet article |
| Blebbistatin | Bleb | Blebbistatin is a widely used muscle and non-muscle myosin II-specific inhibitor that block contractile activity. Blebbistatin shows selectivity and high affinity for multiple class II myosins. Blebbistatin is commonly employed in respirometric experiments with permeabilized muscle fibers (pfi). Permeabilized muscle fibers are sensitive to low oxygen supply due to diffusion restrictions that limit mitochondrial respiration at the core of the fiber bundle. Therefore, hyperoxic conditions are required to counteract this limitation. Further studies have shown that the addition of blebbistatin in the respiration medium prevents fiber contraction, reduces the oxygen sensitivity and allows the study of ADP kinetics in pfi at normoxic oxygen levels. However, other studies described that the presence of blebbistatin does not prevent the oxygen dependence in pfi. Moreover, several limitations of blebbistatin i.e. low solubility in water, cytotoxicity and phototoxicity have been described. |
| Body fat excess | BFE | In the healthy reference population (HRP), there is zero body fat excess, BFE, and the fraction of excess body fat in the HRP is expressed - by definition - relative to the reference body mass, M°, at any given height. Importantly, body fat excess, BFE, and body mass excess, BME, are linearly related, which is not the case for the body mass index, BMI. |
| Body mass | m [kg]; M [kg·x-1] | The body mass M is the mass (kilogram [kg]) of an individual (object) [x] and is expressed in units [kg/x]. Whereas the body weight changes as a function of gravitational force (you are weightless at zero gravity; your floating weight in water is different from your weight in air), your mass is independent of gravitational force, and it is the same in air and water. |
| Body mass excess | BME | The body mass excess, BME, is an index of obesity and as such BME is a lifestyle metric. The BME is a measure of the extent to which your actual body mass, M [kg/x], deviates from M° [kg/x], which is the reference body mass [kg] per individual [x] without excess body fat in the healthy reference population, HRP. A balanced BME is BME° = 0.0 with a band width of -0.1 towards underweight and +0.2 towards overweight. The BME is linearly related to the body fat excess. |
| Body mass index | BMI | The body mass index, BMI, is the ratio of body mass to height squared (BMI=M·H-2), recommended by the WHO as a general indicator of underweight (BMI<18.5 kg·m-2), overweight (BMI>25 kg·m-2) and obesity (BMI>30 kg·m-2). Keys et al (1972; see 2014) emphasized that 'the prime criterion must be the relative independence of the index from height'. It is exactly the dependence of the BMI on height - from children to adults, women to men, Caucasians to Asians -, which requires adjustments of BMI-cutoff points. This deficiency is resolved by the body mass excess relative to the healthy reference population. |
| CDGSH iron-sulfur domain proteins | CISD proteins | The CDGSH iron-sulfur domain (CISDs) family of proteins uniquely ligate labile 2Fe-2S clusters with a 3Cys-1His motif. CISD1 and CISD3 have been demonstrated to localize to the outer mitochondrial membrane and mitochondrial matrix respectively, however their relationship to mitochondrial physiology remains ill-defined [1]. The best characterized member of the CISD family, CISD1, has been demonstrated to be involved in respiratory capacity, iron homeostasis, and ROS regulation |
| Carrier control titrations | Most of the nonpolar compounds have to be diluted in organic solvents such as DMSO or acetonitrile in order to use them for the titrations in the SUIT protocols. However, the solvent (carrier) itself could affect the mitochondrial physiology and promote alterations that we need to take into account. For this reason, it is necessary to run in parallel to our treatment experiment a control experiment on which we will add a carrier control titration to test if it affects our sample or not. | |
| Cell ergometry | ||
| Cell respiration | Cell respiration channels metabolic fuels into the chemiosmotic coupling (bioenergetic) machinery of oxidative phosphorylation, being regulated by and regulating oxygen consumption (or consumption of an alternative final electron acceptor) and molecular redox states, ion gradients, mitochondrial (or microbial) membrane potential, the phosphorylation state of the ATP system, and heat dissipation in response to intrinsic and extrinsic energy demands. See also respirometry. In internal or cell respiration in contrast to fermentation, redox balance is maintained by external electron acceptors, transported into the cell from the environment. The chemical potential between electron donors and electron acceptors drives the electron transfer pathway, generating a chemiosmotic potential that in turn drives ATP synthesis. | |
| Comorbidity | Comorbidities are common in obesogenic lifestyle-induced early aging. These are preventable, non-communicable diseases with strong associations to obesity. In many studies, cause and effect in the sequence of onset of comorbidities remain elusive. Chronic degenerative diseases are commonly obesity-induced. The search for the link between obesity and the etiology of diverse preventable diseases lead to the hypothesis, that mitochondrial dysfunction is the common mechanism, summarized in the term 'mitObesity'. | |
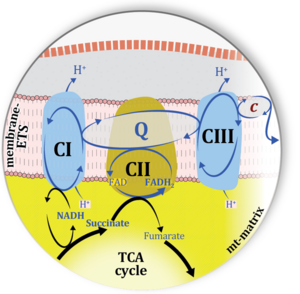
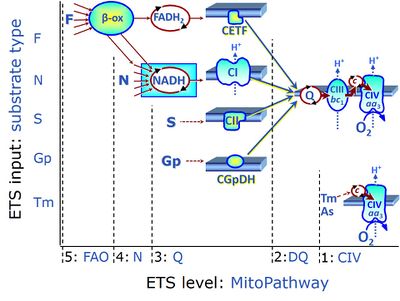
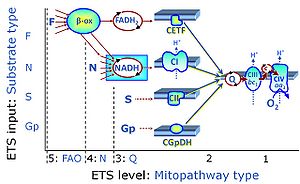
| Complex II ambiguities | CII ambiguities | The current narrative that the reduced coenzymes NADH and FADH2 feed electrons from the tricarboxylic acid (TCA) cycle into the mitochondrial electron transfer system can create ambiguities around respiratory Complex CII. Succinate dehydrogenase or CII reduces FAD to FADH2 in the canonical forward TCA cycle. However, some graphical representations of the membrane-bound electron transfer system (ETS) depict CII as the site of oxidation of FADH2. This leads to the false believe that FADH2 generated by electron transferring flavoprotein (CETF) in fatty acid oxidation and mitochondrial glycerophosphate dehydrogenase (CGpDH) feeds electrons into the ETS through CII. In reality, NADH and succinate produced in the TCA cycle are the substrates of Complexes CI and CII, respectively, and the reduced flavin groups FMNH2 and FADH2 are downstream products of CI and CII, respectively, carrying electrons from CI and CII into the Q-junction. Similarly, CETF and CGpDH feed electrons into the Q-junction but not through CII. The ambiguities surrounding Complex II in the literature call for quality control, to secure scientific standards in current communications on bioenergetics and support adequate clinical applications. |
| Concentration | c [mol·L-1]; C [x·L-1] | Concentration [mol·L-1] is a volume-specific quantity for diluted samples s. In a concentration, the sample is expressed in a variety of formats: count, amount, charge, mass, energy. In solution chemistry, amount concentration is amount of substance nB per volume V of the solution, cB = [B] = nB·V-1 [mol·dm-3] = [mol·L-1]. The standard concentration, c°, is defined as 1 mol·L-1 = 1 M. Count concentration CX = NX·V-1 [x·L-1] is the concentration of the number NX of elementary entities X, for which the less appropriate term 'number concentration' is used by IUPAC. If the sample is expressed as volume Vs (e.g., VO2), then the 'volume-concentration' of Vs in V is termed 'volume fraction', Φs = Vs·V-1 (e.g., volume fraction of O2 in dry air, ΦO2) = 0.20946). Density is the mass concentration in a volume VS of pure sample S. A change of concentration, dcX, in isolated or closed systems at constant volume is due to internal transformations (advancement per volume) only. In closed compressible systems (with a gas phase), the concentration of the gas changes, when pressure-volume work is performed on the system. In open systems, a change of concentration can additionally be due to external flow across the system boundaries. |
| Coupled respiration | Coupled respiration drives oxidative phosphorylation of the diphosphate ADP to the triphosphate ATP, mediated by proton pumps across the inner mitochondrial membrane. Intrinsically uncoupled respiration, in contrast, does not lead to phosphorylation of ADP, despite of protons being pumped across the inner mt-membrane. Coupled respiration, therefore, is the coupled part of respiratory oxygen flux that pumps the fraction of protons across the inner mt-membrane which is utilized by the phosphorylation system to produce ATP from ADP and Pi. In the OXPHOS state, mitochondria are in a partially coupled state, and the corresponding coupled respiration is the free OXPHOS capacity. In the state of ROUTINE respiration, coupled respiration is the free ROUTINE activity. | |
| Coupling-control state | CCS | Coupling-control states are defined in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, homogenates) as LEAK respiration, OXPHOS, and ET states, with corresponding respiration rates (L, P, E) in any electron-transfer-pathway state which is competent for electron transfer. These coupling states are induced by titration of ADP and uncouplers, and application of specific inhibitors of the phosphorylation pathway. In living cells, the coupling-control states are LEAK respiration, ROUTINE, and ET states of respiration with corresponding rates L, R, E, using membrane-permeable inhibitors of the phosphorylation system (e.g. oligomycin) and uncouplers (e.g. CCCP). Coupling-control protocols induce these coupling-control states sequentially at a constant electron-transfer-pathway state. |
| Coupling/pathway control diagram | CPCD | Coupling/pathway control diagrams illustrate the respiratory states obtained step-by-step in substrate-uncoupler-inhibitor titrations in a SUIT protocol. Each step (to the next state) is defined by an initial state and a metabolic control variable, X. The respiratory states are shown by boxes. X is usually the titrated substance in a SUIT protocol. If X (ADP, uncouplers, or inhibitors of the phosphorylation system, e.g. oligomycin) exerts coupling control, then a transition is induced between two coupling-control states. If X (fuel substrates, e.g. pyruvate and succinate, or Electron transfer pathway inhibitors, e.g. rotenone) exerts pathway control, then a transition is induced between two Electron-transfer-pathway states. The type of metabolic control (X) is shown by arrows linking two respiratory states, with vertical arrows indicating coupling control, and horizontal arrows indicating pathway control. Marks define the section of an experimental trace in a given respiratory state (steady state). Events define the titration of X inducing a transition in the SUIT protocol. The specific sequence of coupling control and pathway control steps defines the SUIT protocol pattern. The coupling/pathway control diagrams define the categories of SUIT protocols (see Figure). |
| Crabtree effect | The Crabtree effect describes the observation that respiration is frequently inhibited when high concentrations of glucose or fructose are added to the culture medium - a phenomenon observed in numerous cell types, particularly in proliferating cells, not only tumor cells but also bacteria and yeast. The Pasteur effect (suppression of glycolysis by oxygen) is the converse of the Crabtree effect (suppression of respiration by high concentration of glucose or fructose). | |
| Critical oxygen pressure | pc | The critical oxygen pressure, pc, is defined as the partial oxygen pressure, pO2, below which aerobic catabolism (respiration or oxygen consumption) declines significantly. If anaerobic catabolism is activated simultaneously to compensate for lower aerobic ATP generation, then the limiting oxygen pressure, pl, is equal to the pc. In many cases, however, the pl is substantially lower than the pc. |
| Curcumin | Curcumin has been shown to possess significant anti-inflammatory, anti-oxidant, anti-carcinogenic, anti-mutagenic, anti-coagulant and anti-infective effects. The protective effects of curcumin on rat heart mitochondrial injuries induced by in vitro anoxia–reoxygenation were evaluated by Xu et al 2013. It was found that curcumin added before anoxia or immediately prior to reoxygenation exhibited remarkable protective effects against anoxia–reoxygenation induced oxidative damage to mitochondria. | |
| DatLab and SUIT protocols | This is a brief summary of steps to be taken for performing a high-resolution respirometry experiment with SUIT protocols using the OROBOROS Oroboros O2k and DatLab software. (1) Search for a specific SUIT protocol name (go to MitoPedia: SUIT). The list of MitoPedia SUIT protocols can be sorted by categories of SUIT protocols (sorting by SUIT protocol name), which is listed as the 'abbreviation' of the SUIT protocol name. (2) Copy the template for Mark names into your DatLab subdirectory: DatLab\APPDATA\MTEMPLAT. (3) Copy the DatLab-Analysis template for this SUIT protocol. (4) Follow the link to the corresponding publication or MiPNet communication, where the pdf file describing the SUIT protocol is available. (5) A DatLab demo file may be available providing an experimental example. After each sequential titration, a mark is set on the plot for flux or flow. After having set all marks, pull down the 'Mark names' menu, select the corresponding SUIT protocol for mark names, and rename all marks. The Mark names template also provides standard values of the titration volume preceding each mark. (6) Go to 'Mark statistics' [F2], copy to clipboard, and paste into the sample tab in the DatLab-Analysis template.
| |
| Diapause | Diapause is a preprogrammed form of developmental arrest that allows animals to survive harsh environmental conditions and may also allow populations to synchronize periods of growth and reproduction with periods of optimal temperatures and adequate water and food. Diapause is endogenously controlled, and this dormancy typically begins well before conditions become too harsh to support normal growth and development [1,2]. » MiPNet article | |
| ET capacity | E | |
| Elamipretide | Bendavia | Bendavia (Elamipretide) was developed as a mitochondria-targeted drug against degenerative diseases, including cardiac ischemia-reperfusion injury. Clinical trials showed variable results. It is a cationic tetrapeptide which readily passes cell membranes, associates with cardiolipin in the mitochondrial inner membrane. Supercomplex-associated CIV activity significantly improved in response to elamipretide treatment in the failing human heart. |
| Electron leak | Electrons that escape the electron transfer pathway without completing the reduction of oxygen to water at cytochrome c oxidase, causing the production of ROS. The rate of electron leak depends on the topology of the complex, the redox state of the moiety responsible of electron leakiness and usually on the protonmotive force (Δp). In some cases, the Δp dependance relies more on the ∆pH component than in the ∆Ψ. | |
| Electron transfer pathway | ET pathway | In the mitochondrial electron transfer pathway (ET pathway) electrons are transferred from externally supplied reduced fuel substrates to oxygen. Based on this experimentally oriented definition (see ET capacity), the ET pathway consists of (1) the membrane-bound ET pathway with respiratory complexes located in the inner mt-membrane, (2) TCA cycle and other mt-matrix dehydrogenases generating NADH and succinate, and (3) the carriers involved in metabolite transport across the mt-membranes. » MiPNet article |
| Electron-transfer-pathway state | ET-pathway state |
Electron-transfer-pathway states are obtained in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, tissue homogenate) by depletion of endogenous substrates and addition to the mitochondrial respiration medium of fuel substrates (CHNO) activating specific mitochondrial pathways, and possibly inhibitors of specific pathways. Mitochondrial electron-transfer-pathway states have to be defined complementary to mitochondrial coupling-control states. Coupling-control states require ET-pathway competent states, including oxygen supply. Categories of SUIT protocols are defined according to mitochondrial ET-pathway states. » MiPNet article |
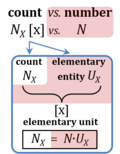
| Elementary unit | x | The elementary unit [x] is the unit of a count NX [x]. The International System of Units defines the unit of a count as 1. Then the Number 1 is the Unit of the Count of Entities — NUCE. This causes a number of formal inconsistencies which are resolved by introducing the elementary unit [x] as the abstracted unit of Euclid’s unit, which is an elementary entity UX [x], and as the unit of Euclid’s number, which is a count NX [x]. |
| Energy charge | AEC | The energy charge of the adenylate system or adenylate energy charge (AEC) has been defined by Atkinson and Walton (1967) as (ATP + ½ ADP)/(AMP + ADP + ATP). Wheather the AEC is a fundamental metabolic control parameter remains a controversial topic. |
| Ergodynamic efficiency | ε | The ergodynamic efficiency, ε (compare thermodynamic efficiency), is a power ratio between the output power and the (negative) input power of an energetically coupled process. Since power [W] is the product of a flow and the conjugated thermodynamic force, the ergodynamic efficiency is the product of an output/input flow ratio and the corresponding force ratio. The efficiency is 0.0 in a fully uncoupled system (zero output flow) or at level flow (zero output force). The maximum efficiency of 1.0 can be reached only in a fully (mechanistically) coupled system at the limit of zero flow at ergodynamic equilibrium. The ergodynamic efficiency of coupling between ATP production (DT phosphorylation) and oxygen consumption is the flux ratio of DT phosphorylation flux and oxygen flux (P»/O2 ratio) multiplied by the corresponding force ratio. Compare with the OXPHOS-coupling efficiency. |
| Ergodynamics | The mission of ergodynamics is the revelation of relations of general validity. "Thermodynamics deals with relationships between properties of systems at equilibrium and with differences in properties between various equilibrium states. It has nothing to do with time. Even so, it is one of the most powerful tools of physical chemistry" [1]. Ergodynamics is the theory of exergy changes (from the Greek word 'erg' which means work). Ergodynamics includes the fundamental aspects of thermodynamics ('heat') and the thermodynamics of irreversible processes (TIP; nonequilibrium thermodynamics), and thus links thermodynamics to kinetics. In its most general scope, ergodynamics is the science of energy transformations. Classical thermodynamics includes open systems, yet as a main focus it describes closed systems. This is reflected in a nomenclature that is not easily applicable to the more general case of open systems [2]. At present, IUPAC recommendations [3] fall short of providing adequate guidelines for describing energy transformations in open systems. | |
| Extensive quantity | Extensive quantities pertain to a total system, e.g. oxygen flow. An extensive quantity increases proportional with system size. The magnitude of an extensive quantity is completely additive for non-interacting subsystems, such as mass or flow expressed per defined system. The magnitude of these quantities depends on the extent or size of the system (Cohen et al 2008). | |
| External flow | Ie [MU·s-1] | External flows across the system boundaries are formally reversible. Their irreversible facet is accounted for internally as transformations in a heterogenous system (internal flows, Ii). |
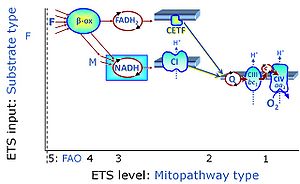
| F-junction | The F-junction is a junction for convergent electron flow in the electron transfer pathway (ET-pathway) from fatty acids through fatty acyl CoA dehydrogenase (reduced form FADH2) to electron transferring flavoprotein (CETF), and further transfer through the Q-junction to Complex III (CIII). The concept of the F-junction and N-junction provides a basis for defining categories of SUIT protocols. Fatty acid oxidation, in the F-pathway control state, not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the N-junction substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCoA into the TCA cycle catalyzed by citrate synthase. | |
| Flavonoids | Flavonoids are a group of bioactive polyphenols with potential antioxidant and anti-inflammatory effects, abundant in fruits and vegetables, and in some medicinal herbs. Flavonoids are synthesized in plants from phenylalanine. Dietary intake of flavonoids as nutraceuticals is discussed for targeting T2D and other degenerative diseases. | |
| Flow | I [MU∙s-1] | In an isomorphic analysis, any form of flow, I is the advancement of a process per unit of time, expressed in a specific motive unit [MU∙s-1], e.g., ampere for electric flow or current [A≡C∙s-1], watt for heat flow [W≡J∙s-1], and for chemical flow the unit is [mol∙s-1]. Flow is an extensive quantity. The corresponding isomorphic forces are the partial exergy (Gibbs energy) changes per advancement [J∙MU-1], expressed in volt for electric force [V≡J∙C-1], dimensionless for thermal force, and for chemical force the unit is [J∙mol-1], which deserves a specific acronym ([Jol]) comparable to volt. |
| Flux | J | Flux, J, is a specific quantity. Flux is flow, I [MU·s-1 per system] (an extensive quantity), divided by system size. Flux (e.g., oxygen flux) may be volume-specific (flow per volume [MU·s-1·L-1]), mass-specific (flow per mass [MU·s-1·kg-1]), or marker-specific (e.g. flow per mtEU). The motive unit [MU] of chemical flow or flux is the advancement of reaction [mol] in the chemical format. |
| Flux control efficiency | jZ-Y | Flux control efficiencies express the control of respiration by a metabolic control variable, X, as a fractional change of flux from YX to ZX, normalized for ZX. ZX is the reference state with high (stimulated or un-inhibited) flux; YX is the background state at low flux, upon which X acts.
Complementary to the concept of flux control ratios and analogous to elasticities of metabolic control analysis, the flux control efficiency of X upon background YX is expressed as the change of flux from YX to ZX normalized for the reference state ZX. » MiPNet article |
| Force | F; dmFX; ΔtrFX [J·MU-1] | Force is an intensive quantity. The product of force times advancement is the work (exergy) expended in a process or transformation. Force times flow is power [W].
|
| Free activity | αX [MU·m-3] | Free activity αX [MU·m-3] is pressure divided by isomorphic force. In the chemical amount format, αX is expressed in units of concentration of X [mol·L-1]. αX is the local concentration in a concentration gradient. If the concentration gradient is collapsed to a boundary of zero thickness in a compartmental system, αX reflects the singularity in the transition between the two phases or compartments. |
| Harmonized SUIT protocols | H-SUIT | Harmonized SUIT protocols (H-SUIT) are designed to include cross-linked respiratory states. When performing harmonized SUIT protocols in parallel, measurements of cross-linked respiratory states can be statistically evaluated as replicates across protocols. Additional information is obtained on respiratory coupling and substrate control by including respiratory states that are not common (not cross-linked) across the harmonized protocols. |
| Healthy ageing | Healthy ageing: 'WHO has released the first World report on ageing and health, reviewing current knowledge and gaps and providing a public health framework for action. The report is built around a redefinition of healthy ageing that centres on the notion of functional ability: the combination of the intrinsic capacity of the individual, relevant environmental characteristics, and the interactions between the individual and these characteristics' (Beard 2016 The Lancet). | |
| Healthy reference population | HRP | A healthy reference population, HRP, establishes the baseline for the relation between body mass and height in healthy people of zero underweight or overweight, providing a reference for evaluation of deviations towards underweight or overweight and obesity. The WHO Child Growth Standards (WHO-CGS) on height and body mass refer to healthy girls and boys from Brazil, Ghana, India, Norway, Oman and the USA. The Committee on Biological Handbooks compiled data on height and body mass of healthy males from infancy to old age (USA), published before emergence of the fast-food and soft-drink epidemic. Four allometric phases are distinguished with distinct allometric exponents. At heights above 1.26 m/x the allometric exponent is 2.9, equal in women and men, and significantly different from the exponent of 2.0 implicated in the body mass index, BMI [kg/m2]. |
| Height of humans | h [m]; H [m·x-1] | The height of humans, h, is given in SI units in meters [m]. Humans are countable objects, and the symbol and unit of the number of objects is N [x]. The average height of N objects is, H = h/N [m/x], where h is the heights of all N objects measured on top of each other. Therefore, the height per human has the unit [m·x-1] (compare body mass [kg·x-1]). Without further identifyer, H is considered as the standing height of a human, measured without shoes, hair ornaments and heavy outer garments. |
| Hydrogen ion pump | Mitochondrial hydrogen ion pumps — frequently referred to as "proton pumps" — are large enzyme complexes (CI, CIII, CIV, ATP synthase) spanning the mt-inner membrane mtIM, partially encoded by mtDNA. CI, CIII and CIV are H+ pumps that drive hydrogen ions against the electrochemical protonmotive force pmF and thus generating the pmF, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CV) is a H+ pump that utilizes the exergy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
| Hydrogenion flux | JH+ | Volume-specific hydrogenion flux or H+ flux is measured in a closed system as the time derivative of H+ concentration, expressed in units [pmol·s-1·mL-1]. H+ flux can be measured in an open system at steady state, when any acidification of the medium is compensated by external supply of an equivalent amount of base. The extracellular acidification rate (ECAR) is the change of pH in the incubation medium over time, which is zero at steady state. Volume-specific H+ flux is comparable to volume-specific oxygen flux [pmol·s-1·mL-1], which is the (negative) time derivative of oxygen concentration measured in a closed system, corrected for instrumental and chemical background. pH is the negative logarithm of hydrogen ion activity. Therefore, ECAR is of interest in relation to acidification issues in the incubation buffer or culture medium. The physiologically relevant metabolic H+ flux, however, must not be confused with ECAR. |
| Hyperoxia | hyperox | Hyperoxia is defined as environmental oxygen pressure above the normoxic reference level. Cellular and intracellular hyperoxia is imposed on isolated cells and isolated mitochondria at air-level oxygen pressures which are higher compared to cellular and intracellular oxygen pressures under tissue conditions in vivo. Hyperoxic conditions may impose oxidative stress and may increase maximum aerobic performance. |
| Hyperthermia | Hyperthermia in endotherms is a state of stressful up to lethal elevated body core temperature. In humans, the limit of hyperthermia (fever) is considered as >38.3 °C, compared to normothermia at a body temperature of 36.5 to 37.5 °C. | |
| Hypothermia | Hypothermia in endotherms is a state of stressful up to lethal low body core temperature. In humans, the limit of hypothermia is considered as 35 °C, compared to normothermia at a body temperature of 36.5 to 37.5 °C. Hypothermia is classified as mild (32–35 °C), moderate (28–32 °C), severe (20–28 °C), and profound (<20 °C). | |
| Hypoxia | hypox | Hypoxia (hypox) is defined in respiratory physiology as the state when insufficient O2 is available for respiration, compared to environmental hypoxia defined as environmental oxygen pressures below the normoxic reference level. Three major categories of hypoxia are (1) environmental hypoxia, (2) physiological tissue hypoxia in hyperactivated states (e.g. at VO2max) with intracellular oxygen demand/supply balance at steady state in tissues at environmental normoxia, compared to tissue normoxia in physiologically balanced states, and (3) pathological tissue hypoxia including ischemia and stroke, anaemia, chronic heart disease, chronic obstructive pulmonary disease, severe COVID-19, and obstructive sleep apnea. Pathological hypoxia leads to tissue hypoxia and heterogenous intracellular anoxia. Clinical oxygen treatment ('environmental hyperoxia') may not or only partially overcome pathological tissue hypoxia. |
| Intensive quantity | Intensive quantities are partial derivatives of an extensive quantity by the advancement, dtrξX, of an energy transformation tr; example: Force. In contrast to extensive quantities which pertain to the entire system and are additive, extensive quantities 'take well defined values at each point of the system' (Prigogine 1967 Interscience) and are non-additive. Intensive and extensive quantities can be easily discriminated by the units, e.g. [J] for the extensive quantity, in contrast to [J·mol-1] for the corresponding intensive quantity. In the general definition of thermodynamics, intensive quantities are not distinguished from specific quantities (Cohen 2008 IUPAC Green Book). Ergodynamics emphasizes the contrast between specific quantities which are extensive quantities normalized for a variable expressing system size (mass, volume of the system, amount of substance in a system) and intensive quantities which are normalized for the motive unit of a transformation (mass exchanged, volume change of the system, amount of substance reacting in a system; Gnaiger 1993 Pure Appl Chem). Intensive and specific quantities are both non-additive, take well defined values at each point of the system, and both corresponding quantities are expressed in identical units, e.g. the intensive quantity Gibbs force of a catabolic reaction (such as oxidation; 0 = -1 Glc - 6 O2 + 6 CO2 + 6 H2O), ΔkGGlc [kJ·mol-1], and the specific quantity Gibbs energy per mole glucose contained in a system, GGlc [kJ·mol-1] (with respect to an arbitrarily defined reference state, such as the reference state of formation or combustion). | |
| Internal flow | Ii [MU·s-1] | Within the system boundaries, irreversible internal flows, Ii,—including chemical reactions and the dissipation of internal gradients of heat and matter—contribute to internal entropy production, diS/dt. In contrast, external flows, Ie, of heat, work, and matter proceed reversibly across the system boundaries (of zero thickness). Flows are expressed in various formats per unit of time, with corresponding motive units [MU], such as chemical [mol], electrical [C], mass [kg]. Flow is an extensive quantity, in contrast to flux as a specific quantity. |
| Jmax | Jmax | Jmax is the maximum pathway flux (e.g. oxygen flux) obtained at saturating substrate concentration. Jmax is a function of metabolic state. In hyperbolic ADP or oxygen kinetics, Jmax is calculated by extrapolation of the hyperbolic function, with good agreement between the calculated and directly measured fluxes, when substrate levels are >20 times the c50 or p50. |
| Limiting oxygen pressure | pl | The limiting oxygen pressure, pl, is defined as the partial oxygen pressure, pO2, below which anaerobic catabolism is activated to contribute to total ATP generation. The limiting oxygen pressure, pl, may be substantially lower than the critical oxygen pressure, pc, below which aerobic catabolism (respiration or oxygen consumption) declines significantly. |
| Matrix-ETS | matrix-ETS | The component of the electron transfer system located in the mitochondrial matrix (matrix-ETS) is distringuished from the ETS bound to the mt-inner membrane (membrane-ETS). Electron transfer and corresponding OXPHOS capacities are classically studied in mitochondrial preparations as oxygen consumption supported by various fuel substrates undergoing partial oxidation in the mt-matrix, such as pyruvate, malate, succinate, and others. |
| Melatonin | aMT | Melatonin (N-acetyl-5-methoxytryptamine, aMT) is a highly conserved molecule present in unicellular to vertebrate organisms. Melatonin is synthesized from tryptophan in the pinealocytes by the pineal gland and also is produced in other organs, tissues and fluids (extrapineal melatonin). Melatonin has lipophilic and hydrophilic nature which allows it to cross biological membranes. Therefore, melatonin is present in all subcellular compartments predominantly in the nucleus and mitochondria. Melatonin has pleiotropic functions with powerful antioxidant, anti-inflammatory and oncostatic effects with a wide spectrum of action particularly at the level of mitochondria. » MiPNet article |
| Metabolic control variable | X | A metabolic control variable X causes the transition between a background state Y (background rate YX) and a reference state Z (reference rate ZX). X may be a stimulator or activator of flux, inducing the step change from background to reference steady state (Y to Z). Alternatively, X may be an inhibitor of flux, absent in the reference state but present in the background state (step change from Z to Y). |
| Metformin | Metformin (dimethylbiguanide) is mainly known as an important antidiabetic drug which is effective, however, in a wide spectrum of degenerative diseases. It is an inhibitor of Complex I and glycerophosphate dehydrogenase complex. | |
| MiPMap | MiPMap |
The project Mitochondrial Physiology Map (MiPMap) is initiated to provide an overview of mitochondrial properties in cell types, tissues and species. As part of Bioblast, MiPMap may be considered as an information synthase for Comparative Mitochondrial Physiology. Establishing a comprehensive database will require global input and cooperation. A comparative database of mitochondrial physiology may provide the key for understanding the functional implications of mitochondrial diversity from mouse to man, and evaluation of altered mitochondrial respiratory control patterns in health and disease (Gnaiger 2009). |
| Microxia | microx | Microxia (deep hypoxia) is obtained when trace amounts of O2 exert a stimulatory effect on respiration above the level where metabolism is switched to a purely anaerobic mode. |
| MitObesity drugs | Bioactive mitObesity compounds are drugs and nutraceuticals with more or less reproducible beneficial effects in the treatment of diverse preventable degenerative diseases implicated in comorbidities linked to obesity, characterized by common mechanisms of action targeting mitochondria. | |
| MitoFit protocols | MitoFit protocols are moderated by the MitoFit moderators (MitoFit team), either as protocols with direct reference to publications available to the scientific communicty, or protocols additionally described and made available in Bioblast with full information on authors (including contact details), author contributions, and editor (moderator) in charge. This aims at a comprehensive MitoFit data repository, which will require global input and cooperation. | |
| Mitochondria | mt | Mitochondria (Greek mitos: thread; chondros: granule) are small structures within cells, which function in cell respiration as powerhouses or batteries. Mitochondria belong to the bioblasts of Richard Altmann. Abbreviation: mt, as generally used in mtDNA. Singular: mitochondrion (bioblast); plural: mitochondria (bioblasts). |
| Mitochondrial competence | mt-competence; MitoCom | Mitochondrial metabolic competence is the organelle's capacity to provide adequate amounts of ATP in due time, by adjusting the mt-membrane potential, mt-redox states and the ATP/ADP ratio according to the metabolic requirements of the cell.
The term mitochondrial competence is also known in a genetic context: Mammalian mitochondria possess a natural competence for DNA import. MitoCom_O2k-Fluorometer is a Mitochondrial Competence network, the nucleus of which is formed by the K-Regio project MitoCom Tyrol. |
| Mitochondrial concentration | CmtE | Mitochondrial concentration is CmtE = mtE·V-1 [mtEU·m-3]. mt-Concentration is an experimental variable, dependent on sample concentration. |
| Mitochondrial content | mtENX | Mitochondrial content per object X is mtENX = mtE·NX-1 [mtEU·x-1]. |
| Mitochondrial density | DmtE | Specific mitochondrial density is DmtE = mtE·mX-1 [mtEU·kg-1]. If the amount of mitochondria, mtE, is expressed as mitochondrial mass, then DmtE is the mass fraction of mitochondria in the sample. If mtE is expressed as mitochondrial volume, Vmt, and the mass of sample, mX, is replaced by volume of sample, VX, then DmtE is the volume fraction of mitochondria in the sample. |
| Mitochondrial inner membrane | mtIM | The mitochondrial inner membrane mtIM is the structure harboring the membrane-bound electron transfer system ETS including the respiratory complexes working as hydrogen ion pumps, the mt-phosphorylation system including the hydrogen ion pump ATP synthase, several substrate transporters involved in the electron transfer pathway, and a variety of other ion pumps that carry proton charge (Ca2+, Mg2+). The protonmotive force is the electrochemical potential difference across the mtIM generated by the hydrogen ion pumps of the . |
| Mitochondrial marker | mt-marker | Mitochondrial markers are structural or functional properties that are specific for mitochondria. A structural mt-marker is the area of the inner mt-membrane or mt-volume determined stereologically, which has its limitations due to different states of swelling. If mt-area is determined by electron microscopy, the statistical challenge has to be met to convert area into a volume. When fluorescent dyes are used as mt-marker, distinction is necessary between mt-membrane potential dependent and independent dyes. mtDNA or cardiolipin content may be considered as a mt-marker. Mitochondrial marker enzymes may be determined as molecular (amount of protein) or functional properties (enzyme activities). Respiratory capacity in a defined respiratory state of a mt-preparation can be considered as a functional mt-marker, in which case respiration in other respiratory states is expressed as flux control ratios. » MiPNet article |
| Mitochondrial matrix | mt-matrix | The mitochondrial matrix (mt-matrix) is enclosed by the mt-inner membrane mtIM. The terms mitochondrial matrix space or mitochondrial lumen are used synonymously. The mt-matrix contains the enzymes of the tricarboxylic acid cycle, fatty acid oxidation and a variety of enzymes that have cytosolic counterparts (e.g. glutamate dehydrogenase, malic enzyme). Metabolite concentrations, such as the concentrations of fuel substrates, adenylates (ATP, ADP, AMP) and redox systems (NADH), can be very different in the mt-matrix, the mt-intermembrane space, and the cytosol. The finestructure of the gel-like mt-matrix is subject of current research. |
| Mitochondrial outer membrane | mtOM | The mitochondrial outer membrane is the incapsulating membrane which is osmotically not active and contains the cytochrome b5 enzyme similar to that found in the endoplasmatic reticulum, the translocases of the outer membrane, monoaminooxidase, the palmitoyl-CoA synthetase and carnytil-CoA transferase 1. |
| Mitochondrial respiration | Integrative measure of the dynamics of complex coupled metabolic pathways, including metabolite transport across the mt-membranes, TCA cycle function with electron transfer through dehydrogenases in the mt-matrix, membrane-bound electron transfer mET-pathway, the transmembrane proton circuit, and the phosphorylation system. | |
| Molar mass | M [kg·mol-1]; [g·mol-1] | Molar mass M is the mass of a chemical compound divided by its amount-of-substance measured in moles. It is defined as MB = m/nB, where m is the total mass of a sample of pure substance and nB is the amount of substance B given in moles. The definition applies to pure substance. The molar mass allows for converting between the mass of a substance and its amount for bulk quantities. It is calculated as the sum of standard atomic weights of all atoms that form one entity of the substance. The appropriate SI base units is kg·mol-1. However, for historical as well as usability reasons, g·mol-1 is almost always used instead. |
| MtOM | mtOM | The mitochondrial outer membrane |
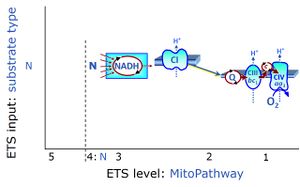
| N-junction | The N-junction is a junction for convergent electron flow in the electron transfer pathway (ET-pathway) from type N substrates (further details »N-pathway control state) through the mt-NADH pool to Complex I (CI), and further transfer through the Q-junction to Complex III (CIII). Representative type N substrates are pyruvate (P), glutamate (G) and malate (M). The corresponding dehydrogenases (PDH, GDH, MDH) and some additional TCA cycle dehydrogenases (isocitrate dehydrogenase, oxoglutarate dehydrogenase generate NADH, the substrate of Complex I (CI). The concept of the N-junction and F-junction provides a basis for defining categories of SUIT protocols based on Electron-transfer-pathway states. | |
| NADH electron transfer-pathway state | N | The NADH electron transfer-pathway state (N) is obtained by addition of NADH-linked substrates (CI-linked), feeding electrons into the N-junction catalyzed by various mt-dehydrogenases. N-supported flux is induced in mt-preparations by the addition of NADH-generating substrate combinations of pyruvate (P), glutamate (G), malate (M), oxaloacetate (Oa), oxoglutarate (Og), citrate, hydroxybutyrate. These N-junction substrates are (indirectly) linked to Complex I by the corresponding dehydrogenase-catalyzed reactions reducing NAD+ to NADH+H+ + H+. The most commonly applied N-junction substrate combinations are: PM, GM, PGM. The malate-anaplerotic pathway control state (M alone) is a special case related to malic enzyme (mtME). The glutamate-anaplerotic pathway control state (G alone) supports respiration through glutamate dehydrogenase (mtGDH). Oxidation of tetrahydrofolate is a NAD(P)H linked pathway with formation of formate. In mt-preparations, succinate dehydrogenase (SDH; CII) is largely substrate-limited in N-linked respiration, due to metabolite depletion into the incubation medium. The residual involvement of S-linked respiration in the N-pathway control state can be further suppressed by the CII-inhibitor malonic acid). In the N-pathway control state ET pathway level 4 is active. |
| NS e-input | NS, CI&II | NS e-input or the NS-pathway control state is electron input from a combination of substrates for the N-pathway control state and S-pathway control state through Complexes CI and CII simultaneously into the Q-junction. NS e-input corresponds to TCA cycle function in vivo, with convergent electron flow through the Electron transfer pathway. In mt-preparations, NS e-input requires addition not only of NADH- (N-) linked substrates (pyruvate&malate or glutamate&malate), but of succinate (S) simultaneously, since metabolite depletion in the absence of succinate prevents a significant stimulation of S-linked respiration. For more details, see: Additive effect of convergent electron flow. |
| Normothermia | Normothermia in endotherms is a state when body core temperature is regulated within standard limits. In humans, normothermia is considered as a body temperature of 36.4 to 37.8 °C. Normothermia, however, has a different definition in the context of ectotherms. » MiPNet article | |
| Normoxia | normox | Normoxia is a reference state, frequently considered as air-level oxygen pressure at sea level (c. 20 kPa in water vapor saturated air) as environmental normoxia. Intracellular tissue normoxia is variable between organisms and tissues, and intracellular oxygen pressure is frequently well below air-level pO2 as a result of cellular (mainly mitochondrial) oxygen consumption and oxygen gradients along the respiratory cascade. Oxygen pressure drops from ambient normoxia of 20 kPa to alveolar normoxia of 13 kPa, while extracellular normoxia may be as low as 1 to 5 kPa in solid organs such as heart, brain, kidney and liver. Pericellular pO2 of cells growing in monolayer cell cultures may be hypoxic compared to tissue normoxia when grown in ambient normoxia (95 % air and 5 % CO2) and a high layer of culture medium causing oxygen diffusion limitation at high respiratory activity, but pericellular pO2 may be effectively hyperoxic in cells with low respiratory rate with a thin layer of culture medium (<2 mm). Intracellular oxygen levels in well-stirred suspended small cells (5 - 7 mm diameter; endothelial cells, fibroblasts) are close to ambient pO2 of the incubation medium, such that matching the experimental intracellular pO2 to the level of intracellular tissue normoxia requires lowering the ambient pO2 of the medium to avoid hyperoxia. |
| Obesity | Obesity is a disease resulting from excessive accumulation of body fat. In common obesity (non-syndromic obesity) excessive body fat is due to an obesogenic lifestyle with lack of physical exercise ('couch') and caloric surplus of food consumption ('potato'), causing several comorbidities which are characterized as preventable non-communicable diseases. Persistent body fat excess associated with deficits of physical activity induces a weight-lifting effect on increasing muscle mass with decreasing mitochondrial capacity. Body fat excess, therefore, correlates with body mass excess up to a critical stage of obesogenic lifestyle-induced sarcopenia, when loss of muscle mass results in further deterioration of physical performance particularly at older age. | |
| Open system | An open system is a system with boundaries that allow external exchange of energy and matter; the surroundings are merely considered as a source or sink for quantities transferred across the system boundaries (external flows, Iext). | |
| Ordinate | y | The ordinate is the vertical axis y of a rectangular two-dimensional graph with the abscissa x as the horizontal axis. Values Y are placed vertically from the origin. See Ordinary Y/X regression. |
| Oxidative stress | Oxidative stress results from an imbalance between pro-oxidants and antioxidants shifting the equilibrium in favor of the pro-oxidants. This process can be due by an increment in pro-oxidants, by a depletion of antioxidant systems or both. Oxidative stress generates oxidative damage of proteins, lipids and DNA. | |
| Oxygen flow | IO2 [mol·s-1] or [mol·s-1·x-1] | Respiratory oxygen flow is the oxygen consumption per total system, which is an extensive quantity. Flow is advancement of a transformation in a system per time [mol·s-1], when 'system' is defined as the experimental system (e.g. an open or closed chamber). Flow is distinguished from the size-specific quantity flux obtained by normalization of flow per volume of the experimental system [mol·s-1·m-3]. An experimental object, e.g. a living cell, may be considered as the 'experimental system'. Then oxygen flow per cell has the unit [mol·s-1·x-1], where [x] is the elementary unit for a count. Oxygen flow or respiration per cell [amol·s-1·x-1] = [pmol·s-1·Mx-1] is normalized for the cell count, distinguished from oxygen flux (e.g. per mg protein or wet mass). These are different forms of normalization of rate. |
| Oxygen flux | JO2 | Oxygen flux, JO2, is a specific quantity. Oxygen flux is oxygen flow, IO2 [mol·s-1 per system] (an extensive quantity), divided by system size. Flux may be volume-specific (flow per volume [pmol·s-1·mL-1]), mass-specific (flow per mass [pmol·s-1·mg-1]), or marker-specific (flow per mtEU). Oxygen flux (e.g., per body mass, or per cell volume) is distinguished from oxygen flow (per number of objects, such as cells), IO2 [mol·s-1·x-1]. These are different forms of normalization of rate. |
| Oxygen flux - instrumental background | J°O2 | Instrumental background oxygen flux, J°O2, in a respirometer is due to oxygen consumption by the POS, and oxygen diffusion into or out of the aqueous medium in the O2k-chamber. It is a property of the instrumental system, measured in the range of experimental oxygen levels by a standardized instrumental O2 background test. The oxygen regime from air saturation towards zero oxygen is applied generally in experiments with isolated mitochondria, and living or permeabilized cells. To overcome oxygen diffusion limitation in permeabilized fibers and homogenates, an elevated oxygen regime is applied, requiring instrumental background test in the same range of elevated oxygen. |
| Oxygen solubility | SO2 [µM/kPa] | The oxygen solubility, SO2 [µM/kPa] = [(µmol·L-1)/kPa], expresses the oxygen concentration in solution in equilibrium with the oxygen pressure in a gas phase, as a function of temperature and composition of the solution. The inverse of oxygen solubility is related to the activity of dissolved oxygen. The oxygen solubility in solution, SO2(aq), depends on temperature and the concentrations of solutes in solution, whereas the dissolved oxygen concentration at equilibrium with air, cO2*(aq), depends on SO2(aq), barometric pressure and temperature. SO2(aq) in pure water is 10.56 µM/kPa at 37 °C and 12.56 µM/kPa at 25 °C. At standard barometric pressure (100 kPa), cO2*(aq) is 207.3 µM at 37 °C (19.6 kPa partial oxygen pressure) or 254.7 µM at 25 °C (20.3 kPa partial oxygen pressure). In MiR05 and serum, the corresponding saturation concentrations are lower due to the oxygen solubility factor: 191 and 184 µM at 37 °C or 234 and 227 µM at 25 °C. |
| P50 | p50 | p50 is the oxygen partial pressure at which (a) respiratory flux is 50% of maximum oxygen flux, Jmax, at saturating oxygen levels. The oxygen affinity is indirectly proportional to the p50. The p50 depends on metabolic state and rate. (b) p50 is the oxygen partial pressure at which oxygen binding (on myoglobin, haemoglobin) is 50%, or desaturation is 50%. |
| PH | pH | The pH value or pH is the negative of the base 10 logarithm of the activity of protons (hydrogen ions, H+). A pH electrode reports the pH and is sensitive to the activity of H+. In dilute solutions, the hydrogen ion activity is approximately equal to the hydrogen ion concentration. The symbol pH stems from the term potentia hydrogenii. |
| Phosphorylation pathway | DT | The phosphorylation pathway (phosphorylation system) is the functional unit utilizing the protonmotive force to phosphorylate ADP (D) to ATP (T), and may be defined more specifically as the P»-system. The P»-system consists of adenine nucleotide translocase, phosphate carrier, and ATP synthase. Mitochondrial adenylate kinase, mt-creatine kinase and mt-hexokinase constitute extended components of the P»-system, controlling local AMP and ADP concentrations and forming metabolic channels. Since substrate-level phosphorylation is involved in the TCA-cycle, the P»-system includes succinyl-CoA ligase (GDP to GTP or ADP to ATP). |
| Physiological pathway-control state | See Electron-transfer-pathway state. | |
| Pressure | p, P, Π [Pa] | Pressure is a fundamental quantity expressing energy per volume. The SI unit of pressure is generally pascal [Pa] = [J·m-3]. The term 'stress' (mechanical stress) is used as a synonym for pressure (SI). Pressure is known in physics as mechanical pressure, which is force per area, p = F·A-1 [Pa] = [N·m-2]. In physical chemistry, gas pressure is defined as p = n·V-1·RT, where the concentration is c = n·V-1 [mol·m-3], R is the gas constant, and T is the absolute temperature, and RT is expressed in units of chemical force [J·mol-1]. van't Hoff's osmotic pressure assumes the same form applied to dissolved substances diffusing across a semipermeable membrane, but concentrations should be replaced by activities. The activity of dissolved gases is expressed by the partial pressure, where the solubility can be seen as an activity coefficient. Pressure appears explicitely or implicitely in all chapters of physics and physical chemistry. In contrast to the universal counterparts energy and force, however, the general connections between various isomorphic expressions of pressure remain poorly understood: Pressure is the concentration of the force at the point of action. More generally, pressure is the force times concentration at the interphase of interaction. |
| Proton pump | Mitochondrial proton pumps are large enzyme complexes (CI, CIII, CIV, CV) spanning the inner mt-membrane, partially encoded by mtDNA. CI, CIII and CIV are proton pumps that drive protons against the electrochemical protonmotive force, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CIV) is a proton pump that utilizes the energy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
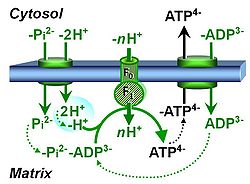
| Protonmotive force | pmF, ∆mFH+, Δp [J·MU-1] | The protonmotive force ∆mFH+ is known as Δp in Peter Mitchell’s chemiosmotic theory [1], which establishes the link between electric and chemical components of energy transformation and coupling in oxidative phosphorylation. The unifying concept of the pmF ranks among the most fundamental theories in biology. As such, it provides the framework for developing a consistent theory and nomenclature for mitochondrial physiology and bioenergetics. The protonmotive force is not a vector force as defined in physics. This conflict is resolved by the generalized formulation of isomorphic, compartmental forces, ∆trF, in energy (exergy) transformations [2]. Protonmotive means that there is a potential for the movement of protons, and force is a measure of the potential for motion.
The pmF is generated in oxidative phosphorylation by oxidation of reduced fuel substrates and reduction of O2 to H2O, driving the coupled proton translocation from the mt-matrix space across the mitochondrial inner membrane (mtIM) through the proton pumps of the electron transfer pathway (ETS), which are known as respiratory Complexes CI, CIII and CIV. ∆mFH+ consists of two partial isomorphic forces: (1) The chemical part, ∆dFH+, relates to the diffusion (d) of uncharged particles and contains the chemical potential difference§ in H+, ∆µH+, which is proportional to the pH difference, ∆pH. (2) The electric part, ∆elFp+ (corresponding numerically to ∆Ψ)§, is the electric potential difference§, which is not specific for H+ and can, therefore, be measured by the distribution of any permeable cation equilibrating between the negative (matrix) and positive (external) compartment. Motion is relative and not absolute (Principle of Galilean Relativity); likewise there is no absolute potential, but isomorphic forces are stoichiometric potential differences§. The total motive force (motive = electric + chemical) is distinguished from the partial components by subscript ‘m’, ∆mFH+. Reading this symbol by starting with the proton, it can be seen as pmF, or the subscript m (motive) can be remembered by the name of Mitchell, ∆mFH+ = ∆dFH+ + ∆elFp+ With classical symbols, this equation contains the Faraday constant, F, multiplied implicitly by the charge number of the proton (zH+ = 1), and has the form [1] ∆p = ∆µH+∙F-1 + ∆ΨA partial electric force of 0.2 V in the electrical format, ∆elFeH+a, is 19 kJ∙mol-1 H+a in the molar format, ∆elFnp+a. For 1 unit of ∆pH, the partial chemical force changes by -5.9 kJ∙mol-1 in the molar format, ∆dFnH+a, and by 0.06 V in the electrical format, ∆dFeH+a. Considering a driving force of -470 kJ∙mol-1 O2 for oxidation, the thermodynamic limit of the H+a/O2 ratio is reached at a value of 470/19 = 24, compared to the mechanistic stoichiometry of 20 for the N-pathway with three coupling sites. |
| Protonmotive pressure | pmP, ∆mΠH+ [kPa] | The protonmotive pressure, ∆mΠH+ or pmP [kPa], is an extension of Peter Mitchell’s concept of the protonmotive force pmF, based on Fick’s law of diffusion and Einstein’s diffusion equation, accounting for osmotic pressure (corresponding to the diffusion term in the pmF) and electric pressure (the electric term or membrane potential in the pmF). The linearity of the generalized flow-pressure relationship explains the non-ohmic flow-force dependence in the proton leak rate as a function of membrane potential.
The total motive pressure (motive = electric + chemical) is distinguished from the partial components by subscript ‘m’, ∆mΠH+, ∆mΠH+ = ∆dΠH+ + ∆elΠp+ |
| P»-system | P»system | The ADP-ATP phosphorylation system or P»-system. See Phosphorylation system. |
| Q | Q | Multiple meanings of Q
|
| Q-junction | The Q-junction is a junction for convergent electron flow in the Electron transfer pathway (ET-pathway) from type N substrates and mt-matrix dehydrogenases through Complex I (CI), from type F substrates and FA oxidation through electron-transferring flavoprotein Complex (CETF), from succinate (S) through Complex II (CII), from glycerophosphate (Gp) through glycerophosphate dehydrogenase Complex (CGpDH), from choline through choline dehydrogenase, from dihydro-orotate through dihydro-orotate dehydrogenase, and other enzyme Complexes into the Q-cycle (ubiquinol/ubiquinone), and further downstream to Complex III (CIII) and Complex IV (CIV). The concept of the Q-junction, with the N-junction and F-junction upstream, provides the rationale for defining Electron-transfer-pathway states and categories of SUIT protocols. | |
| Q-pools | Q | Different Q-pools are more or less clearly distinguished in the cell, related to a variety of models describing degress of Q-pool behavior. (1) CoQ-pools are distinguished according to their compartmentation in the cell: mitochondrial CoQ (mtCoQ) and CoQ in other organelles versus plasma-membrane CoQ. (2) The total mitochondrial CoQ-pool mtCoQ is partitioned into an ETS-reactive Q-pool, Qra, and an inactive mtCoQ-pool, Qia. (2a) The Qra-pool is fully reduced in the form of quinol QH2 under anoxia, and fully oxidized in the form of quinone in aerobic mitochondrial preparations incubated without CHNO-fuel substrates. Intermediate redox states of Qra are sensitive to pathway control and coupling control of mitochondrial electron transfer and OXPHOS. (2b) The Qia-pool remains partially reduced and oxidized independent of aerobic-anoxic transitions. The redox state of Qia is insensitive to changes in mitochondrial respiratory states. (3) The Qra-pool is partitioned into Q with Q-pool behavior according to the fluid-state model (synonymous: random-collision model) and Q tightly bound to supercomplexes according to the solid-state model. The two models describe the extremes in a continuum of homogenous or heterogenous Q-pool behavior. The CII-Q-CIII segment of the S-pathway is frequently considered to follow homogenous Q-pool behavior participating in the Qhom-pool, whereas the CI-Q-CIII segment of the N-pathway indicates supercomplex organization and metabolic channeling with different degrees of Q-pool heterogeneity contributing to the Qhet-pool. |
| RT | RT versus RT | RT indicates room temperature or 25 °C. RT is the gas constant R [kJ/mol] multiplied by absolute temperature T [K]. This is the motive force quantum in the amount format (Gnaiger 2020 BEC MitoPathways). |
| Rapamycin | Rapamycin is an inhibitor of the mammalian/mechanistic target of rapamycin, complex 1 (mTORC1). Rapamycin induces autophagy and dyscouples mitochondrial respiration. Rapamycin delays senescence in human cells, and extends lifespan in mice without detrimental effects on mitochondrial fitness in skeletal muscle. | |
| Research | Research is a term composed of search and re. What does this tell us? The best comparison of the English with a German word is Untersuchung, composed of suchung (search) and unter (below). The term search (suchen) is straightforward to understand and comparable in both languages. The prefix re and unter are more difficult to reconcile, yet in both languages these perfixes reveal complementary if not nearly identical messages. re means {Quote} back to the original place; again, anew, once more {end of Quote} [1], whereas unter means below or underneath. Re-search, therefore, is not simply the search or investigation of some topic or problem, it means essentially doing the search again and again (re -> re-producibility) and penetrating below a simple search by reaching out for an underlying level of the search. The re in re-search and re-producibility has to be extended ultimately from a single re-search group to inter-laboratory re-investigation. This tells us, therefore, that while search is valuable, re-search provides the necessary validation. This re-evaluation of confirmative re-search should be re-cognized as the most important strategy to address the reproducibility crisis. | |
| Respiratory state | Respiratory states of mitochondrial preparations and living cells are defined in the current literature in many ways and with a diversity of terms. Mitochondrial respiratory states must be defined in terms of both, the coupling-control state and the electron-transfer-pathway state. | |
| Respirometry | Respirometry is the quantitative measurement of respiration. Respiration is therefore a combustion, a very slow one to be precise (Lavoisier and Laplace 1783). Thus the basic idea of using calorimetry to explore the sources and dynamics of heat changes were present in the origins of bioenergetics (Gnaiger 1983). Respirometry provides an indirect calorimetric approach to the measurement of metabolic heat changes, by measuring oxygen uptake (and carbon dioxide production and nitrogen excretion in the form of ammonia, urea, or uric acid) and converting the oxygen consumed into an enthalpy change, using the oxycaloric equivalent. Liebig (1842) showed that the substrate of oxidative respiration was protein, carbohydrates, and fat. The sum of these chemical changes of materials under the influence of living cells is known as metabolism (Lusk 1928). The amount (volume STP) of carbon dioxide expired to the amount (volume STP) of oxygen inspired simultaneously is the respiratory quotient, which is 1.0 for the combustion of carbohydrate, but less for lipid and protein. Voit (1901) summarized early respirometric studies carried out by the Munich school on patients and healthy controls, concluding that the metabolism in the body was not proportional to the combustibility of the substances outside the body, but that protein, which burns with difficulty outside, metabolizes with the greatest ease, then carbohydrates, while fats, which readily burns outside, is the most difficultly combustible in the organism. Extending these conclusions on the sources of metabolic heat changes, the corresponding dynamics or respiratory control was summarized (Lusk 1928): The absorption of oxygen does not cause metabolism, but rather the amount of the metabolism determines the amount of oxygen to be absorbed. .. metabolism regulates the respiration. | |
| Resveratrol | Resveratrol is a natural bioactive phenol prouced by several plants with antioxidant and anti-inflammatory effects. Dietary intake as nutraceutical is discussed for targeting mitochondria with a wide spectrum of action in degenerative diseases. | |
| STPD | STPD | At standard temperature and pressure dry (STPD: 0 °C = 273.15 K and 1 atm = 101.325 kPa = 760 mmHg), the molar volume of an ideal gas, Vm, and Vm,O2 is 22.414 and 22.392 L∙mol-1, respectively. Rounded to three decimal places, both values yield the conversion factor of 0.744 from units used in spiroergometry (VO2max [mL O2·min-1]) to SI units [µmol O2·s-1]. For comparison at normal temperature and pressure dry (NTPD: 20 °C), Vm,O2 is 24.038 L∙mol-1. Note that the SI standard pressure is 100 kPa, which corresponds to the standard molar volume of an ideal gas of 22.711 L∙mol-1 and 22.689 L∙mol-1 for O2. |
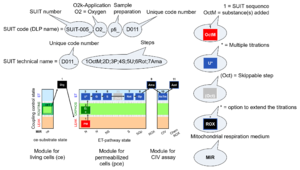
| SUIT | SUIT | SUIT is the abbreviation for Substrate-Uncoupler-Inhibitor Titration. SUIT protocols are used with mt-preparations to study respiratory control in a sequence of coupling and pathway control states induced by multiple titrations within a single experimental assay. These studies use biological samples economically to gain maximum information with a minimum amount of cells or tissue. |
| SUIT protocol library | SUITs | The Substrate-uncoupler-inhibitor titration (SUIT) protocol library contains a sequential list of SUIT protocols (D001, D002, ..) with links to the specific SUIT pages. Classes of SUIT protocols are explained with coupling and substrate control defined for mitochondrial preparations. |
| SUIT protocol pattern | SUITp-Pattern | The SUIT protocol pattern describes the type of the sequence of coupling and substrate control steps in a SUIT protocol, which may be liner, orthogonal, or diametral. |
| SUIT-001 | RP1 | 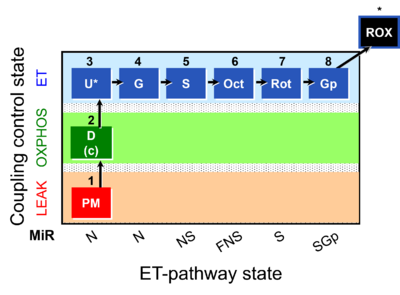 |
| SUIT-002 | RP2 | 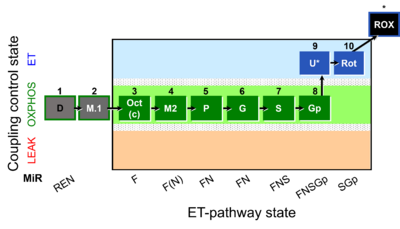 |
| SUIT-003 | CCP-ce | 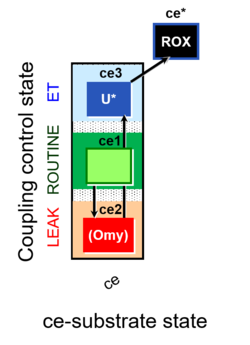 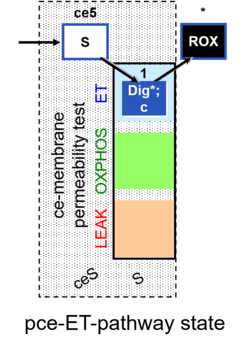 |
| SUIT-004 | RP1-short | 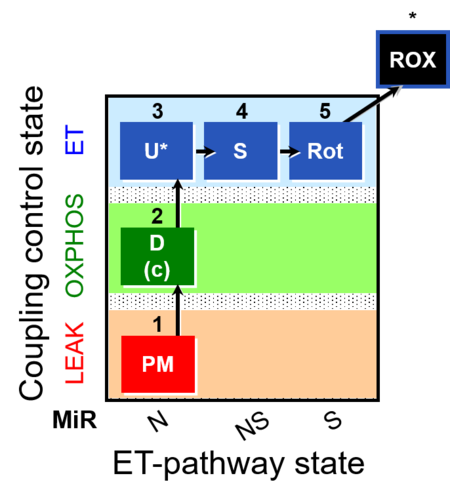 |
| SUIT-005 | RP2-short | 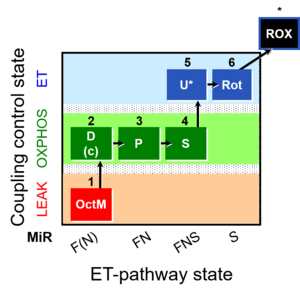 |
| SUIT-006 | CCP-mtprep | 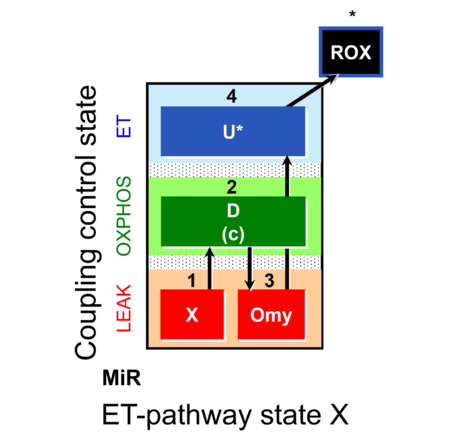 |
| SUIT-007 | Glutamate anaplerosis | 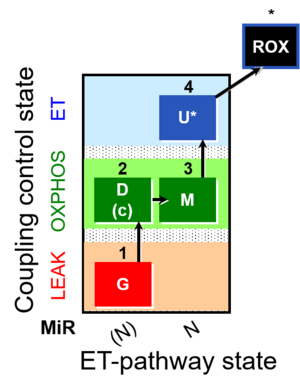 |
| SUIT-008 | PM+G+S_OXPHOS+Rot_ET | 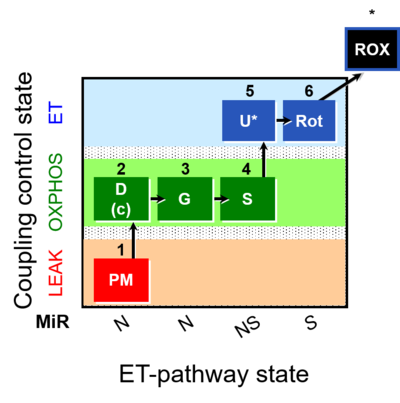 |
| SUIT-009 | 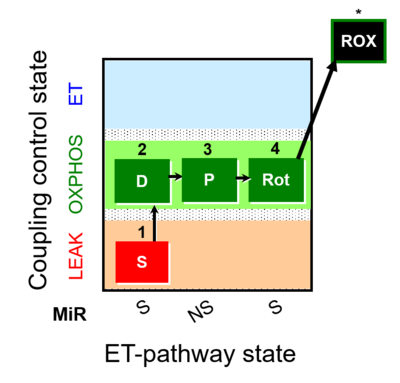 | |
| SUIT-010 | Digitonin test | 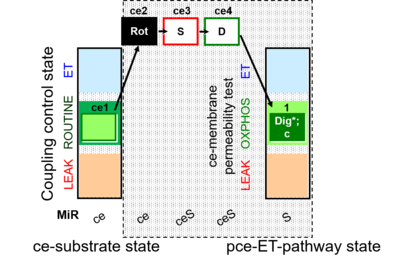 |
| SUIT-011 | GM+S_OXPHOS+Rot_ET | 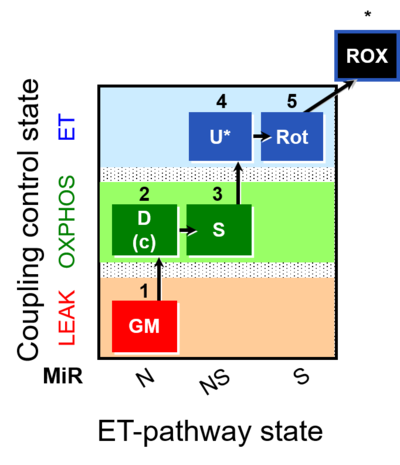 |
| SUIT-012 | PM+G_OXPHOS | 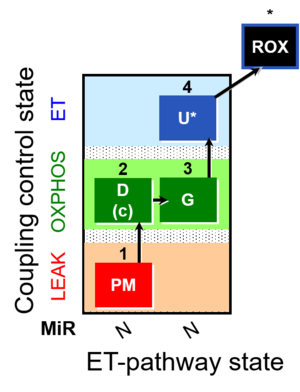 |
| SUIT-013 | ce | 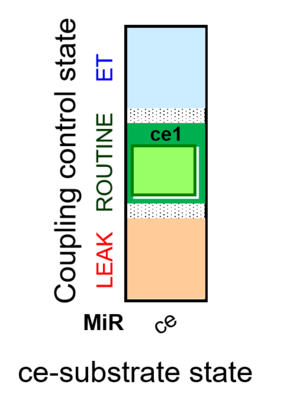 |
| SUIT-014 | GM+P+S_OXPHOS+Rot_ET | 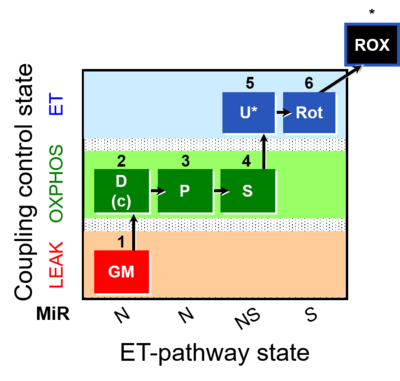 |
| SUIT-015 | F+G+P+S_OXPHOS+Rot_ET | 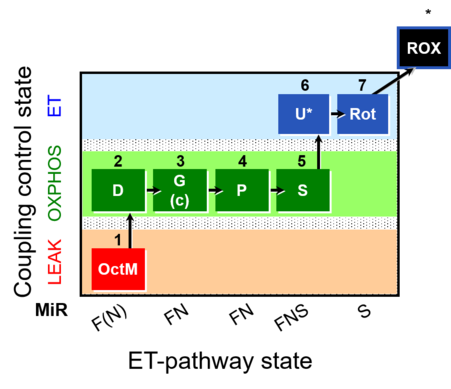 |
| SUIT-016 | F+G+S+Rot_OXPHOS+Omy | 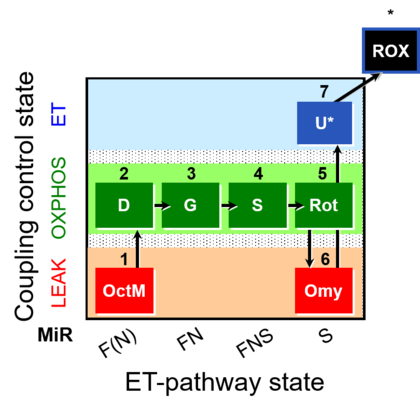 |
| SUIT-017 | F+G+S_OXPHOS+Rot_ET | 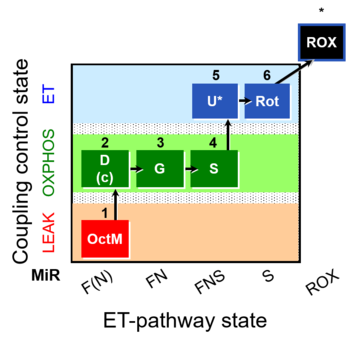 |
| SUIT-018 | O2 dependence-H2O2 | 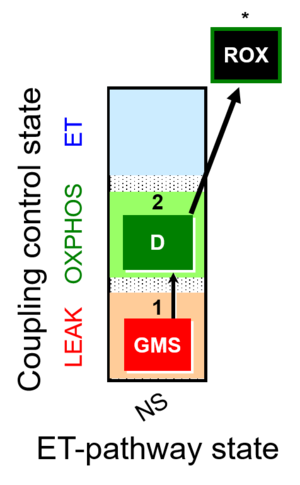 |
| SUIT-019 | Pal+Oct+P+G_OXPHOS+S+Rot_ET | 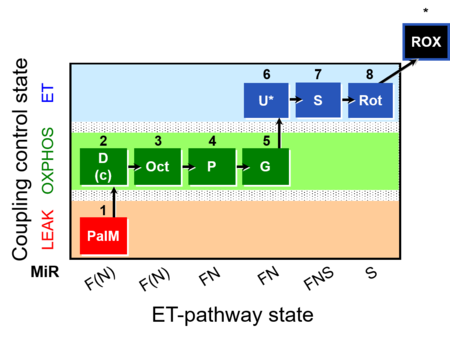 |
| SUIT-020 | PM+G+S+Rot_OXPHOS+Omy | 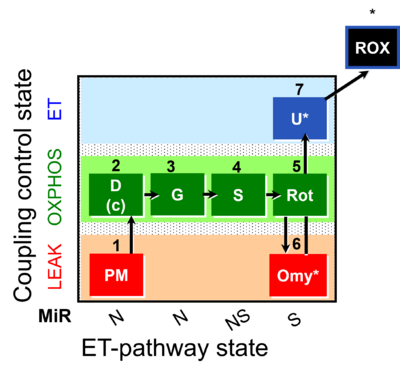 |
| SUIT-021 | OXPHOS (GM+S+Rot+Omy) | 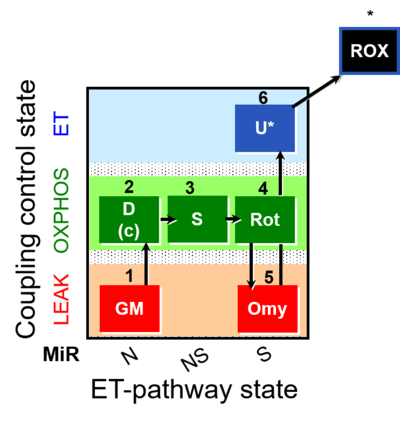 |
| SUIT-022 | AOX (ce CN+SHAM) | 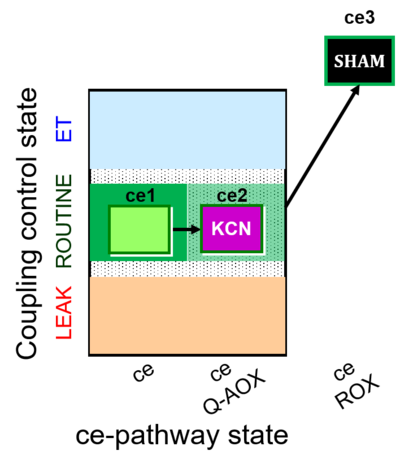 |
| SUIT-024 | ATPase (PM) | 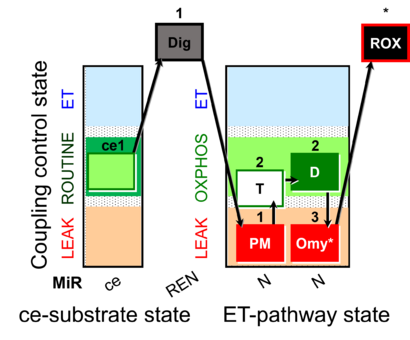 |
| SUIT-025 | OXPHOS (F+M+P+G+S+Rot) | 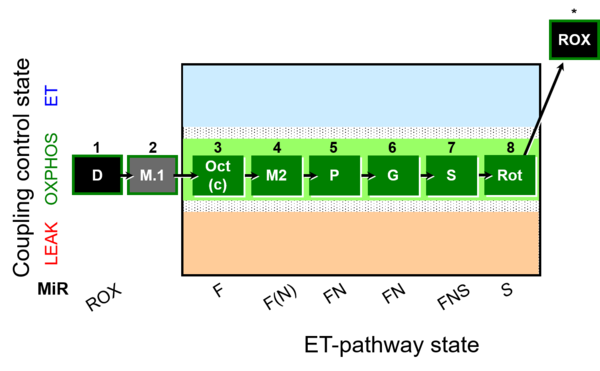 |
| SUIT-027 | Malate anaplerosis | 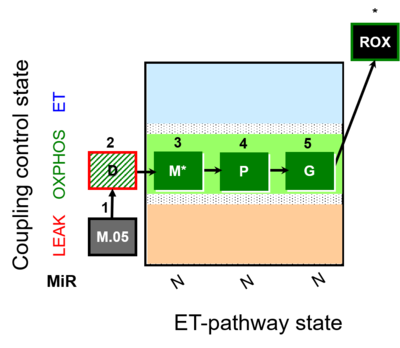 |
| SUIT-031 | PM+S+Rot | 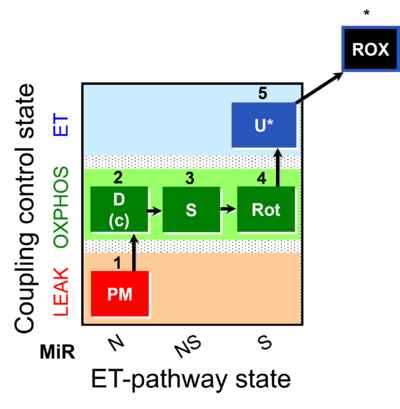 |
| Sample mass concentration | Cms | Sample mass concentration is Cms = ms·V-1 [kg·m-3]. |
| Sarcopenia | Low muscle strength is a key characteristic of sarcopenia due to low muscle quantity and quality, with poor physical performance at severe sarcopenia. Older age may be defined as the age group when sarcopenia becomes a common burden. | |
| Science - the concept | Science | As per the 2017 UNESCO Recommendation on Science and Scientific Researchers, the term ‘science’ signifies the enterprise whereby humankind, acting individually or in small or large groups, makes an organized attempt, in cooperation and in competition, by means of the objective study of observed phenomena and its validation through sharing of findings and data and through peer review, to discover and master the chain of causalities, relations or interactions; brings together in a coordinated form subsystems of knowledge by means of systematic reflection and conceptualization; and thereby furnishes itself with the opportunity of using, to its own advantage, understanding of the processes and phenomena occurring in nature and society. |
| Solubility | SG | The solubility of a gas, SG, is defined as concentration divided by partial pressure, SG = cG·pG-1. |
| Solutions | A solution is {Quote}: A liquid or solid phase containing more than one substance, when for convenience one (or more) substance, which is called the solvent, is treated differently from the other substances, which are called solutes. When, as is often but not necessarily the case, the sum of the mole fractions of solutes is small compared with unity, the solution is called a dilute solution. A superscript attached to the ∞ symbol for a property of a solution denotes the property in the limit of infinite dilution {end of Quote: IUPAC Gold Book}. » MiPNet article | |
| Specific quantity | Specific quantities are obtained when the extensive quantity is divided by system size, in contrast to intensive quantities. The adjective specific before the name of an extensive quantity is often used to mean divided by mass (Cohen et al 2008). A mass-specific quantity (e.g., mass-specific flux is flow divided by mass of the system) is independent of the extent of non-interacting homogenous subsystems. If mass-specific oxygen flux is constant and independent of system size (expressed as mass), then there is no interaction between the subsystems. The well-established scaling law in respiratory physiology reveals a strong interaction of oxygen consumption and body mass by the fact that mass-specific basal metabolic rate (oxygen flux) does not increase proportionally and linearly with body mass. Maximum mass-specific oxygen flux, VO2max, is less mass-dependent across a large range of body mass of different mammalian species (Weibel and Hoppeler 2005). | |
| Spermidine | Spermidine is a polycationic bioactive polyamine mainly found in wheat germ, soybean and various vegetables, involved in the regulation of mitophagy, cell growth and cell death. Like other caloric restriction mimetics, spermidine is effective in cardioprotection, neuroprotection and anticancer immunosuppression by preserving mitochondrial function and control of autophagy. | |
| Substrate control state | See Electron-transfer-pathway state | |
| Substrate-uncoupler-inhibitor titration | SUIT | Mitochondrial Substrate-uncoupler-inhibitor titration (SUIT) protocols are used with mitochondrial preparations to study respiratory control in a sequence of coupling and substrates states induced by multiple titrations within a single experimental assay. |
| Viruses and mitochondrial medicine | Virus-mt-Medicine | Not enough is known about viruses and mitochondrial medicine, although several studies point towards a link between viral infection and mitochondrial dysfunction using high-resolution respirometry, with potential impact on drug development. |
| Volume | V [m3]; 1 m3 = 1000 L | Volume V is a derived quantity based on the SI base quantity length [m] and is expressed in terms of SI base units in the derived unit cubic meter [m3]. The liter [L = dm3] is a conventional unit of volume for concentration and is used for most solution chemical kinetics. The volume V contained in a system (experimental chamber) is separated from the environment by the system boundaries; this is called the volume of the system, and described in practical language as big/small (derived from length, height) or voluminous. Systems are defined at constant volume or constant pressure. For a pure sample S, the volume VS of the pure sample equals the volume V of the system, VS = V. For sample s in a mixture, the ratio Vs·V-1 is the nondimensional volume fraction Φs of sample s. Quantities divided by volume are concentrations of sample s in a mixture, such as count concentration CX = NX·V-1 [x·L-1], and amount of substance concentration CB = nB·V-1 [mol·L-1]. Mass concentration is density ρs = ms·V-1 [kg·L-1]. In closed compressible systems (with a gas phase), the concentration of the gas increases, when pressure-volume work is performed on the system. |
| Volume of the solute | Most of the chemicals for SUIT protocol titrations are prepared by weighing the substance on the balance, transferring to a volumetric glass flask and adding solvent until the intended volume is reached. However, for practical reasons some of the chemical compounds are prepared by just adding the solvent instead of adjusting it's volume. For example, this approach is useful if the substance is very toxic. Then an arbitratry amount is taken, its mass determined on the balance without trying to reach a specific value and the necessary amount of solvent is added. Adding the solvent instead of adjusting its volume is also useful if small amounts are needed (e.g. 1 mL) or if the compound has to be prepared directly before using it like Pyruvate. In these cases the volume contributed by the solute was tested. |