Risiglione 2020 Int J Mol Sci
| Risiglione P, Leggio L, Cubisino SAM, Reina S, Paternò G, Marchetti B, Magrì A, Iraci N, Messina A (2020) High-resolution respirometry reveals MPP+ mitochondrial toxicity mechanism in a cellular model of parkinson's disease. Int J Mol Sci 21:E7809. https://doi.org/10.3390/ijms21217809 |
Risiglione Pierpaolo, Leggio Loredana, Cubisino Salvatore A M, Reina Simona, Paterno Greta, Marchetti Bianca, Magri Andrea, Iraci Nunzio, Messina Angela (2020) Int J Mol Sci
Abstract: MPP+ is the active metabolite of MPTP, a molecule structurally similar to the herbicide Paraquat, known to injure the dopaminergic neurons of the nigrostriatal system in Parkinson's disease models. Within the cells, MPP+ accumulates in mitochondria where it inhibits complex I of the electron transport chain, resulting in ATP depletion and neuronal impairment/death. So far, MPP+ is recognized as a valuable tool to mimic dopaminergic degeneration in various cell lines. However, despite a large number of studies, a detailed characterization of mitochondrial respiration in neuronal cells upon MPP+ treatment is still missing. By using high-resolution respirometry, we deeply investigated oxygen consumption related to each respiratory state in differentiated neuroblastoma cells exposed to the neurotoxin. Our results indicated the presence of extended mitochondrial damage at the inner membrane level, supported by increased LEAK respiration, and a drastic drop in oxygen flow devoted to ADP phosphorylation in respirometry measurements. Furthermore, prior to complex I inhibition, an enhancement of complex II activity was observed, suggesting the occurrence of some compensatory effect. Overall our findings provide a mechanistic insight on the mitochondrial toxicity mediated by MPP+, relevant for the standardization of studies that employ this neurotoxin as a disease model. • Keywords: MPP+, Parkinson’s disease, SH-SY5Y cells, High-resolution respirometry, Mitochondria • Bioblast editor: Plangger M • O2k-Network Lab: IT Catania Messina A
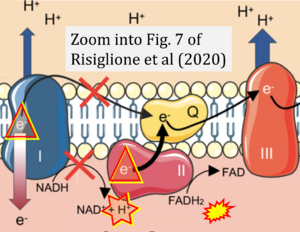
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
On terminology
- For harmonization of terminology on respiratory states and rates, see
- Gnaiger E et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. Bioenerg Commun 2020.1. https://doi.org/10.26124/bec:2020-0001.v1
- For harmonization of terminology on respiratory states and rates, see
Labels:
MitoEAGLE terminology
Labels: MiParea: Respiration, Pharmacology;toxicology
Organism: Human
Tissue;cell: Neuroblastoma
Preparation: Permeabilized cells, Intact cells
Enzyme: Complex II;succinate dehydrogenase
Coupling state: LEAK, ROUTINE, OXPHOS, ET Pathway: N, S, NS, ROX HRR: Oxygraph-2k
2020-11