|
|
| Line 1: |
Line 1: |
| {{MitoPedia | | {{MitoPedia |
| |abbr=CII ambiguities | | |abbr=CII ambiguities |
| |description=[[File:CII-ambiguities Graphical abstract.png|300px|left]] | | |description=[[File:CII-ambiguities Graphical abstract.png|300px|left|link=Gnaiger 2023 MitoFit CII]] |
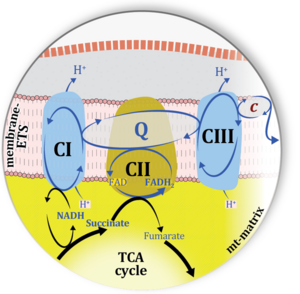
| Two ambiguities or misconceptions around respiratory Complex II (CII) have their roots in the narrative that reduced coenzymes (NADH and FADH<sub>2</sub>) feed electrons from the tricarboxylic acid (TCA) cycle into the mitochondrial electron transfer system. In graphical representations propagating the first ambiguity, succinate dehydrogenase or CII in the canonical (forward) TCA cycle is shown to reduce FAD to FADH<sub>2</sub> (correct), yet CII in the membrane-bound electron transfer system (ETS) is paradoxically represented as the site of oxidation of FADH<sub>2</sub> to FAD. With minor expansion of the tale on electron transfer from FADH<sub>2</sub> into CII, we arrive at the misconception that FADH<sub>2</sub> generated by electron transferring flavoprotein (CETF) in fatty acid oxidation and by mitochondrial glycerophosphate dehydrogenase (CGpDH) feeds electrons into the ETS through CII. For clarification, recall that NADH and succinate formed in the TCA cycle in the mitochondrial matrix are the upstream substrates of Complexes CI and CII, whereas the reduced flavin groups FMNH<sub>2</sub> of flavin mononucleotide and FADH<sub>2</sub> of flavin adenine dinucleotide are products of CI and CII, respectively, with downstream electron flow from CI and CII into the [[Q-junction]]. CETF and CGpDH feed electrons into the Q-junction convergent with but not through CII into the Q-junction. Numerous Complex II ambiguities in the literature with discrepancies in graphical representations and text require quality control to secure scientific standards in current communications on bioenergetics.
| | The current narrative that the reduced coenzymes NADH and FADH2 feed electrons from the tricarboxylic acid (TCA) cycle into the mitochondrial electron transfer system can create ambiguities around respiratory Complex CII. Succinate dehydrogenase or CII reduces FAD to FADH2 in the canonical forward TCA cycle. However, some graphical representations of the membrane-bound electron transfer system (ETS) depict CII as the site of oxidation of FADH2. This leads to the false believe that FADH2 generated by electron transferring flavoprotein (CETF) in fatty acid oxidation and mitochondrial glycerophosphate dehydrogenase (CGpDH) feeds electrons into the ETS through CII. In reality, NADH and succinate produced in the TCA cycle are the substrates of Complexes CI and CII, respectively, and the reduced flavin groups FMNH2 and FADH2 are downstream products of CI and CII, respectively, carrying electrons from CI and CII into the Q-junction. Similarly, CETF and CGpDH feed electrons into the Q-junction but not through CII. The ambiguities surrounding Complex II in the literature call for quality control, to secure scientific standards in current communications on bioenergetics and support adequate clinical applications. - naiger E (2023) Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system. MitoFit Preprints 2023.3. https://doi.org/10.26124/mitofit:2023-0003 |
| }} | | }} |
| __TOC__
| |
| Communicated by [[Gnaiger E]] (2023-03-03) last update 2023-03-24
| |
| == Is there a problem ? ==
| |
| :::::: [[File:Arnold, Finley 2022 Fig1.png|600px|link=Arnold 2023 J Biol Chem]]
| |
| :::: Ref. [1] Arnold PK, Finley LWS (2023) Regulation and function of the mammalian tricarboxylic acid cycle. J Biol Chem 299:102838. - [[Arnold 2023 J Biol Chem |»Bioblast link«]]
| |
| <br>
| |
|
| |
|
| |
| == FADH<sub>2</sub> and FMNH<sub>2</sub> in the S- and N-pathways ==
| |
| [[File:N-S FADH2-FMNH2.png|right|400px]]
| |
| :::: Respiratory Complex CII participates both in the membrane-bound electron transfer system (membrane-ETS) and TCA cycle (matrix-ETS plus CII; [[BEC_2020.1_doi10.26124bec2020-0001.v1 |Gnaiger et al 2020]]). Branches of electron transfer from the reduced coenzyme NADH of nicotinamide adenine dinucleotide N and succinate S converge at the Q-junction in the ETS ('''Figure ;a''' modified from [[Gnaiger_2020_BEC_MitoPathways |Gnaiger 2020]]).
| |
|
| |
| :::: The reduced flavin groups FADH<sub>2</sub> of flavin adenine dinucleotide and FMNH<sub>2</sub> of flavin mononucleotide are at functionally comparable levels in the electron transfer to Q from CII and CI, respectively, just as succinate and NADH are the comparable reduced substrates of CII and CI, respectively ([[Gnaiger_2020_BEC_MitoPathways |Gnaiger 2020]]). In CII the oxidized form FAD is reduced by succinate to the product FADH<sub>2</sub> and the oxidized product fumarate in the TCA cycle. In CI FMN is reduced by NADH forming FMNH<sub>2</sub> and the oxidized NAD<sup>+</sup>. FADH<sub>2</sub> and FMNH<sub>2</sub> are reoxidized downstream in CII and CI by electron transfer to Q in the membrane-bound ETS ('''Figure b''').
| |
|
| |
| :::: Ref. [2] Gnaiger E (2020) Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis. 5th ed. https://doi.org/10.26124/bec:2020-0002
| |
| :::: Ref. [3] Gnaiger E et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. https://doi.org/10.26124/bec:2020-0001.v1
| |
|
| |
|
| |
| === The source and consequence of Complex II ambiguities ===
| |
|
| |
| :::: Ambiguities emerge if the presentation of a concept is vague to an extent that allows for equivocal interpretations. As a consequence of ambiguous representations, even a basically clear and quite simple concept may be communicated further without appropriate reflection as an erroneous divergence from an established truth. The following quotes from Cooper (2000) provide an example.
| |
|
| |
| :::::: [[File:Cooper 2000 Sunderland 10-9.png|700px]]
| |
| :::: Ref. [4] Cooper GM (2000) The cell: a molecular approach. 2nd edition. Sunderland (MA): Sinauer Associates Available from: https://www.ncbi.nlm.nih.gov/books/NBK9885/ - [[Cooper 2000 Sunderland (MA): Sinauer Associates |»Bioblast link«]]
| |
| :::: (''1'') 'Electrons from NADH enter the electron transport chain in complex I, .. A distinct protein complex (complex II), which consists of four polypeptides, receives electrons from the citric acid cycle intermediate, succinate (Figure 10.9). These electrons are transferred to FADH<sub>2</sub>, rather than to NADH, and then to coenzyme Q.'
| |
| :::::: ''Comment'': Here, the frequent comparison is made between FADH<sub>2</sub> (linked to CII) and NADH (linked to CI).
| |
|
| |
| :::: (''2'') 'In contrast to the transfer of electrons from NADH to coenzyme Q at complex I, the transfer of electrons from FADH<sub>2</sub> to coenzyme Q is not associated with a significant decrease in free energy and, therefore, is not coupled to ATP synthesis.'
| |
| :::::: ''Comment'': Note that CI is '''''in''''' the path of the transfer of electrons from NADH to coenzyme Q. In contrast, the transfer of electrons from FADH<sub>2</sub> to coenzyme Q is '''''downstream''''' of CII. Thus even a large Gibbs force ('decrease in free energy') in FADH<sub>2</sub>→Q would fail to drive the coupled process of proton translocation through CII, since the Gibbs force in S→FADH<sub>2</sub> is missing. (In parentheses: None of these steps are coupled to ATP synthesis. Redox-driven proton translocation should not be confused with ''pmF''-driven phosphorylation of ADP).
| |
|
| |
| :::: (''3'') 'Electrons from succinate enter the electron transport chain via FADH<sub>2</sub> in complex II. They are then transferred to coenzyme Q and carried through the rest of the electron transport chain ..'
| |
| :::: ''Comment'': The ambiguity is caused by a lack of unequivocal definition of the electron transfer system ('electron transport chain'). CII receives electrons (''1'') from succinate, yet it is suggested that electrons (from succinate) enter the electron transport chain (''3'') via FADH<sub>2</sub> in complex II. Then two contrasting definitions are implied of the term 'electron transport chain' or better membrane-bound electron transfer system, membrane-ETS. (''a'') If CII is part of the membrane-ETS, then electrons enter the membrane-ETS from succinate (''1'') but not from FADH<sub>2</sub>. (''b'') If electrons enter the 'electron transport chain' via FADH<sub>2</sub> in Complex II (''3''), then CII would be upstream and hence not part of the membrane-ETS (to which conclusion, obviously - see '''Figure''' - nobody would agree). Dismissing concept (''b'') of the membrane-ETS, then remains the ambiguity, if electrons enter the membrane-ETS from FADH<sub>2</sub> (''3'', wrong) or from succinate (''1'', correct).
| |
|
| |
| === FADH<sub>2</sub> - FAD confusion in the S-pathway ===
| |
|
| |
| :::: FADH<sub>2</sub> appears in several publications as the substrate of CII in the electron transfer system linked to succinate oxidation. It is surprising that this error is widely propagated particularly in the most recent literature. For clarification, see [[Gnaiger_2020_BEC_MitoPathways |Gnaiger (2020) page 48]].
| |
|
| |
| :::: The following examples are listed chronologically and illustrate
| |
| ::::'''(1)''' '''ambiguities''' in graphical representations: '''FADH<sub>2</sub> is the product and substrate of CII''' in the same figure, e.g. DeBerardinis, Chandel (2016);
| |
| ::::'''(2)''' '''evolving errors''' in graphical representations: e.g. from Figure 6 (ambiguity) to Figure 1 (error) in Chandel (2021);
| |
| ::::'''(3)''' ambiguities with '''discrepancies between graphical representation and text''': e.g. Figure 1 (error) and text in Fisher-Wellman, Neufer (2012) - 'Reducing equivalents (NADH, FADH<sub>2</sub>) provide electrons that flow through complex I, the ubiquinone cycle (Q/QH<sub>2</sub>), complex III, cytochrome ''c'', complex IV, and to the final acceptor O<sub>2</sub> to form water' (correct);
| |
| ::::'''(4)''' simple '''graphical errors''': e.g. Brownlee (2001), Yépez et al (2018), Chen et al (2022); to
| |
| ::::'''(5)''' propagation of the '''error in the graphical representation solidified by text''': e.g. Arnold, Finley (2022) with the following quotes:
| |
| ::::::* 'SDH reduces FAD to FADH<sub>2</sub>, which donates its electrons to complex II';
| |
| ::::::* 'each complete turn of the TCA cycle generates three NADH and one FADH<sub>2</sub> molecules, which donate their electrons to complex I and complex II, respectively';
| |
| ::::::* 'complex I and complex II oxidize NADH and FADH<sub>2</sub>, respectively'.
| |
|
| |
| :::::: [[File:Arnold, Finley 2022 CORRECTION.png|600px|link=Arnold 2023 J Biol Chem]]
| |
| :::: Ref. [1] Arnold PK, Finley LWS (2023) Regulation and function of the mammalian tricarboxylic acid cycle. J Biol Chem 299:102838. - [[Arnold 2023 J Biol Chem |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Chen 2022 Am J Physiol Cell Physiol CORRECTION.png|500px|link=Chen 2022 Am J Physiol Cell Physiol]]
| |
| :::: Ref. [5] Chen CL, Zhang L, Jin Z, Kasumov T, Chen YR (2022) Mitochondrial redox regulation and myocardial ischemia-reperfusion injury. Am J Physiol Cell Physiol 322:C12-23. - [[Chen 2022 Am J Physiol Cell Physiol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Turton 2022 Int J Mol Sci CORRECTION.png|500px|link=Turton 2022 Int J Mol Sci]]
| |
| :::: Ref. [6] Turton N, Cufflin N, Dewsbury M, Fitzpatrick O, Islam R, Watler LL, McPartland C, Whitelaw S, Connor C, Morris C, Fang J, Gartland O, Holt L, Hargreaves IP (2022) The biochemical assessment of mitochondrial respiratory chain disorders. Int J Mol Sci 23:7487. - [[Turton 2022 Int J Mol Sci |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Ahmad 2022 StatPearls CORRECTION.png|400px|link=Ahmad 2022 StatPearls Publishing]]
| |
| :::: Ref. [7] Ahmad M, Wolberg A, Kahwaji CI (2022) Biochemistry, electron transport chain. StatPearls Publishing StatPearls [Internet]. Treasure Island (FL) - [[Ahmad 2022 StatPearls Publishing |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Yuan 2022 Oxid Med Cell Longev CORRECTION.png|500px|link=Yuan 2022 Oxid Med Cell Longev]]
| |
| :::: Ref. [8] Yuan Q, Zeng ZL, Yang S, Li A, Zu X, Liu J (2022) Mitochondrial stress in metabolic inflammation: modest benefits and full losses. Oxid Med Cell Longev 2022:8803404. - [[Yuan 2022 Oxid Med Cell Longev |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::: [[File:Chandel 2021 Cold Spring Harb Perspect Biol CORRECTION.png|1000px|link=Chandel 2021 Cold Spring Harb Perspect Biol]]
| |
| :::: Ref. [9] Chandel NS (2021) Mitochondria. Cold Spring Harb Perspect Biol 13:a040543. - [[Chandel 2021 Cold Spring Harb Perspect Biol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Yin 2021 FASEB J CORRECTION.png|500px|link=Yin 2021 FASEB J]]
| |
| :::: Ref. [10] Yin M, O'Neill LAJ (2021) The role of the electron transport chain in immunity. FASEB J 35:e21974. - [[Yin 2021 FASEB J |»Bioblast link«]]
| |
|
| |
| :::::: [[File:Missaglia 2021 CORRECTION.png|500px|link=Missaglia 2021 Crit Rev Biochem Mol Biol]]
| |
| :::: Ref. [11] Missaglia S, Tavian D, Angelini C (2021) ETF dehydrogenase advances in molecular genetics and impact on treatment. Crit Rev Biochem Mol Biol 56:360-72. - [[Missaglia 2021 Crit Rev Biochem Mol Biol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Gasmi 2021 Arch Toxicol CORRECTION.png|500px|link=Gasmi 2021 Arch Toxicol]]
| |
| :::: Ref. [12] Gasmi A, Peana M, Arshad M, Butnariu M, Menzel A, Bjørklund G (2021) Krebs cycle: activators, inhibitors and their roles in the modulation of carcinogenesis. Arch Toxicol 95:1161-78. - [[Gasmi 2021 Arch Toxicol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Turton 2021 Expert Opinion Orphan Drugs CORRECTION.png|400px|link=Turton 2021 Expert Opinion Orphan Drugs]]
| |
| :::: Ref. [13] Turton N, Bowers N, Khajeh S, Hargreaves IP, Heaton RA (2021) Coenzyme Q10 and the exclusive club of diseases that show a limited response to treatment. Expert Opinion on Orphan Drugs 9:151-60. - [[Turton 2021 Expert Opinion Orphan Drugs |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Martinez-Reyes, Chandel 2020 CORRECTION.png|600px|link=Martinez-Reyes 2020 Nat Commun]]
| |
| :::: Ref. [14] Martínez-Reyes I, Chandel NS (2020) Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 11:102. - [[Martinez-Reyes 2020 Nat Commun |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Raimondi 2020 Br J Cancer CORRECTION.png|400px|link=Raimondi 2020 Br J Cancer]]
| |
| :::: Ref. [15] Raimondi V, Ciccarese F, Ciminale V (2020) Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br J Cancer 122:168-81. - [[Raimondi 2020 Br J Cancer |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Morelli 2019 Open Biol CORRECTION.png|500px|link=Morelli 2019 Open Biol]]
| |
| :::: Ref. [16] Morelli AM, Ravera S, Calzia D, Panfoli I (2019) An update of the chemiosmotic theory as suggested by possible proton currents inside the coupling membrane. Open Biol 9:180221. - [[Morelli 2019 Open Biol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Lewis 2019 CORRECTION.png|500px|link=Lewis 2019 Int J Mol Sci]]
| |
| :::: Ref. [17] Lewis MT, Kasper JD, Bazil JN, Frisbee JC, Wiseman RW (2019) Quantification of mitochondrial oxidative phosphorylation in metabolic disease: application to Type 2 diabetes. Int J Mol Sci 20:5271. - [[Lewis 2019 Int J Mol Sci |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Sarmah 2019 Transl Stroke Res CORRECTION.png|500px|link=Sarmah 2019 Transl Stroke Res]]
| |
| :::: Ref. [18] Sarmah D, Kaur H, Saraf J, Vats K, Pravalika K, Wanve M, Kalia K, Borah A, Kumar A, Wang X, Yavagal DR, Dave KR, Bhattacharya P (2019) Mitochondrial dysfunction in stroke: implications of stem cell therapy. Transl Stroke Res doi: 10.1007/s12975-018-0642-y - [[Sarmah 2019 Transl Stroke Res |»Bioblast link«]]
| |
|
| |
| :::::: [[File:Yepez 2018 PLOS One Fig1B.jpg|300px|link=Yepez 2018 PLOS One]]
| |
| :::: Ref. [19] Yépez VA, Kremer LS, Iuso A, Gusic M, Kopajtich R, Koňaříková E, Nadel A, Wachutka L, Prokisch H, Gagneur J (2018) OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. PLOS ONE 13:e0199938. - [[Yepez 2018 PLOS One |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Chowdhury 2018 Oxid Med Cell Longev CORRECTION.png|400px|link=Chowdhury 2018 Oxid Med Cell Longev]]
| |
| :::: Ref. [20] Roy Chowdhury S, Banerji V (2018) Targeting mitochondrial bioenergetics as a therapeutic strategy for chronic lymphocytic leukemia. Oxid Med Cell Longev 2018:2426712. - [[Chowdhury 2018 Oxid Med Cell Longev |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:De Villiers 2018 Adv Exp Med Biol CORRECTION.png|400px|link=De Villiers 2018 Adv Exp Med Biol]]
| |
| :::: Ref. [21] de Villiers D, Potgieter M, Ambele MA, Adam L, Durandt C, Pepper MS (2018) The role of reactive oxygen species in adipogenic differentiation. Adv Exp Med Biol 1083:125-144. - [[De Villiers 2018 Adv Exp Med Biol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Zhang 2018 Mil Med Res CORRECTION.png|400px|link=Zhang 2018 Mil Med Res]]
| |
| :::: Ref. [22] Zhang H, Feng YW, Yao YM (2018) Potential therapy strategy: targeting mitochondrial dysfunction in sepsis. Mil Med Res 5:41. - [[Zhang 2018 Mil Med Res |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Polyzos 2017 Mech Ageing Dev CORRECTION.png|400px|link=Polyzos 2017 Mech Ageing Dev]]
| |
| :::: Ref. [23] Polyzos AA, McMurray CT (2017) The chicken or the egg: mitochondrial dysfunction as a cause or consequence of toxicity in Huntington's disease. Mech Ageing Dev 161:181-97. - [[Polyzos 2017 Mech Ageing Dev |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Jones, Bennett 2017 Chapter 4 CORRECTION.png|400px|link=Jones 2017 Elsevier]]
| |
| :::: Ref. [24] Jones PM, Bennett MJ (2017) Chapter 4 - Disorders of mitochondrial fatty acid β-oxidation. Elsevier In: Garg U, Smith LD , eds. Biomarkers in inborn errors of metabolism. Clinical aspects and laboratory determination:87-101. - [[Jones 2017 Elsevier |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:DeBerardinis, Chandel 2016 CORRECTION.png|600px|link=DeBerardinis 2016 Sci Adv]]
| |
| :::: Ref. [25] DeBerardinis RJ, Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2:e1600200. - [[DeBerardinis 2016 Sci Adv |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Nsiah-Sefaa 2016 Bioscie Reports CORRECTION.png|600px|link=Nsiah-Sefaa 2016 Biosci Rep]]
| |
| :::: Ref. [26] Nsiah-Sefaa A, McKenzie M (2016) Combined defects in oxidative phosphorylation and fatty acid β-oxidation in mitochondrial disease. Biosci Rep 36:e00313. - [[Nsiah-Sefaa 2016 Biosci Rep |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Prochaska 2013 Springer CORRECTION.png|400px|link=Prochaska 2013 Springer]]
| |
| :::: Ref. [27] Prochaska LJ, Cvetkov TL (2013) Mitochondrial electron transport. In: Roberts, G.C.K. (eds) Encyclopedia of Biophysics. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-16712-6_25 - [[Prochaska 2013 Springer |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Fisher-Wellman 2012 Trends Endocrinol Metab CORRECTION.png|400px|link=Fisher-Wellman 2012 Trends Endocrinol Metab]] [[File:Fisher-Wellman 2012 Trends Endocrinol Metab Fig2 CORRECTION.png|400px|link=Fisher-Wellman 2012 Trends Endocrinol Metab]]
| |
| :::: Ref. [28] Fisher-Wellman KH, Neufer PD (2012) Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab 23:142-53. - [[Fisher-Wellman 2012 Trends Endocrinol Metab |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Benard 2011 Springer CORRECTION.png|500px|link=Benard 2011 Springer]]
| |
| :::: Ref. [29] Benard G, Bellance N, Jose C, Rossignol R (2011) Relationships between mitochondrial dynamics and bioenergetics. In: Lu Bingwei (ed) Mitochondrial dynamics and neurodegeneration. Springer ISBN 978-94-007-1290-4:47-68. - [[Benard 2011 Springer |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Nussbaum 2005 J Clin Invest CORRECTION.png|500px|link=Nussbaum 2005 J Clin Invest]]
| |
| :::: Ref. [30] Nussbaum RL (2005) Mining yeast in silico unearths a golden nugget for mitochondrial biology. J Clin Invest 115:2689-91. - [[Nussbaum 2005 J Clin Invest |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Sanchez et al 2001 CORRECTION.png|600px|link=Sanchez 2001 Br J Pharmacol]]
| |
| :::: Ref. [31] Sanchez H, Zoll J, Bigard X, Veksler V, Mettauer B, Lampert E, Lonsdorfer J, Ventura-Clapier R (2001) Effect of cyclosporin A and its vehicle on cardiac and skeletal muscle mitochondria: relationship to efficacy of the respiratory chain. Br J Pharmacol 133:781-8. - [[Sanchez 2001 Br J Pharmacol |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Himms-Hagen, Harper 2001 CORRECTION.png|250px|link=Himms-Hagen 2001 Exp Biol Med (Maywood)]]
| |
| :::: Ref. [32] Himms-Hagen J, Harper ME (2001) Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp Biol Med (Maywood) 226:78-84. - [[Himms-Hagen 2001 Exp Biol Med (Maywood) |»Bioblast link«]]
| |
| <br>
| |
|
| |
| :::::: [[File:Brownlee 2001 Nature CORRECTION.png|400px|link=Brownlee 2001 Nature]]
| |
| :::: Ref. [33] Brownlee M (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 14:813-20. - [[Brownlee 2001 Nature |»Bioblast link«]]
| |
| :::: Ref. [34] Arden GB, Ramsey DJ (2015) Diabetic retinopathy and a novel treatment based on the biophysics of rod photoreceptors and dark adaptation. Webvision In: Kolb H, Fernandez E, Nelson R, eds. Webvision: The organization of the retina and visual system [Internet]. Salt Lake City (UT): University of Utah Health Sciences Center; 1995-. - [[Arden 2015 Webvision |»Bioblast link«]]
| |
| <br>
| |
|
| |
| === FADH<sub>2</sub>→CII misconceptions: Websites ===
| |
|
| |
| :::: The following graphs show zooms into the CII-related sections of figures found on the websites cited below. Erroneous presentations are marked by symbols.
| |
|
| |
| :::::: [[File:OpenStax Biology.png|400px]]
| |
| :::: '''Website 1''': [https://openstax.org/books/biology/pages/7-4-oxidative-phosphorylation OpenStax Biology] - Fig. 7.10 Oxidative phosphorylation (CC BY 3.0). - OpenStax Biology got it wrong in figures and text. The error is copied without quality assessment and propagated in several links.
| |
| :::: '''Website 2''': [https://opentextbc.ca/biology/chapter/4-3-citric-acid-cycle-and-oxidative-phosphorylation/ Concepts of Biology] - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19a
| |
| :::: '''Website 3''': [https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_General_Biology_(Boundless)/07%3A_Cellular_Respiration/7.11%3A_Oxidative_Phosphorylation_-_Electron_Transport_Chain LibreTexts Biology] - Figure 7.11.1
| |
| :::: '''Website 4''': [https://courses.lumenlearning.com/wm-biology1/chapter/reading-electron-transport-chain/ lumen Biology for Majors I] - Fig. 1
| |
| :::: '''Website 5''': [https://www.pharmaguideline.com/2022/01/electron-transport-chain.html Pharmaguideline]
| |
|
| |
| :::::: [[File:Khan Academy modified from OpenStax CORRECTION.png|300px]]
| |
| :::: '''Website 6''': [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy] - Image modified from "Oxidative phosphorylation: Figure 1", by OpenStax College, Biology (CC BY 3.0). Figure and text underscore the FADH<sub>2</sub>-error: "''FADH<sub>2</sub> .. feeds them ''(electrons)'' into the transport chain through complex II.''"
| |
| :::: '''Website 7''': [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy]
| |
|
| |
| :::::: [[File:Jack Westin CORRECTION.png|400px]]
| |
| :::: '''Website 8''': [https://jackwestin.com/resources/mcat-content/oxidative-phosphorylation/electron-transfer-in-mitochondria Jack Westin MCAT Courses]
| |
|
| |
| :::::: [[File:Expii OpenStax CORRECTION.png|300px]]
| |
| :::: '''Website 1''': [https://openstax.org/books/biology/pages/7-4-oxidative-phosphorylation OpenStax Biology] - Fig. 7.12
| |
| :::: '''Website 6''': [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy] - Image modified from "Oxidative phosphorylation: Figure 3," by Openstax College, Biology (CC BY 3.0)
| |
| :::: '''Website 7''': [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy]
| |
| :::: '''Website 9''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Image source: By CNX OpenStax
| |
|
| |
| :::::: [[File:Labxchange CORRECTION.png|400px]]
| |
| :::: '''Website 10''': [https://www.labxchange.org/library/items/lb:LabXchange:005ad47f-7556-3887-b4a6-66e74198fbcf:html:1 Labxchange] - Figure 8.15 credit: modification of work by Klaus Hoffmeier
| |
|
| |
| :::::: [[File:Biologydictionary.net CORRECTION.png|400px]]
| |
| :::: '''Website 4''': [https://courses.lumenlearning.com/wm-biology1/chapter/reading-electron-transport-chain/ lumen Biology for Majors I] - Fig. 3
| |
| :::: '''Website 9''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - By OpenStax College CC BY 3.0, via Wikimedia Commons
| |
| :::: '''Website 11''': [https://commons.wikimedia.org/w/index.php?curid=30148497 wikimedia 30148497 - Anatomy & Physiology, Connexions Web site. http://cnx.org/content/col11496/1.6/, 2013-06-19]
| |
| :::: '''Website 12''': [https://biologydictionary.net/electron-transport-chain-and-oxidative-phosphorylation/ biologydictionary.net 2018-08-21]
| |
| :::: '''Website 13''': [https://www.quora.com/Why-does-FADH2-form-2-ATP Quora]
| |
| :::: '''Website 14''': [https://teachmephysiology.com/biochemistry/atp-production/electron-transport-chain/ TeachMePhysiology] - Fig. 1. 2023-03-13
| |
| :::: '''Website 15''': [https://www.thoughtco.com/electron-transport-chain-and-energy-production-4136143 ThoughtCo]
| |
| :::: '''Website 16''': [https://www.toppr.com/ask/question/short-long-answer-types-whatis-the-electron-transport-system-and-what-are-its-functions/ toppr]
| |
|
| |
| :::::: [[File:Researchtweet CORRECTION.png|400px]]
| |
| :::: '''Website 17''': [https://researchtweet.com/mitochondrial-electron-transport-chain-2/ researchtweet]
| |
| :::: '''Website 18''': [https://microbenotes.com/electron-transport-chain/ Microbe Notes]
| |
|
| |
| :::::: [[File:BiochemDen CORRECTION.png|400px]]
| |
| :::: '''Website 19''': [https://biochemden.com/electron-transport-chain-mechanism/ BiochemDen.com]
| |
|
| |
| :::::: [[File:Vector Mine CORRECTION.png|400px]]
| |
| :::: '''Website 20''': [https://www.dreamstime.com/electron-transport-chain-as-respiratory-embedded-transporters-outline-diagram-electron-transport-chain-as-respiratory-embedded-image235345232 dreamstime]
| |
| :::: '''Website 21''': [https://vectormine.com/item/electron-transport-chain-as-respiratory-embedded-transporters-outline-diagram/ VectorMine]
| |
|
| |
| :::::: [[File:Creative-biolabs CORRECTION.png|400px]]
| |
| :::: '''Website 22''': [https://www.creative-biolabs.com/drug-discovery/therapeutics/electron-transport-chain.htm creative-biolabs]
| |
|
| |
| :::::: [[File:Khan Academy CORRECTION.png|400px]]
| |
| :::: '''Website 6''': [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy]
| |
| :::: '''Website 7''': [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy]
| |
|
| |
| :::::: [[File:Expii-Whitney, Rolfes 2002 CORRECTION.png|400px]]
| |
| :::: '''Website 9''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Whitney, Rolfes 2002
| |
|
| |
| :::::: [[File:FlexBooks 2 0 CORRECTION.png|400px]]
| |
| :::: '''Website 23''': [https://flexbooks.ck12.org/cbook/ck-12-biology-flexbook-2.0/section/2.28/primary/lesson/electron-transport-bio/ FlexBooks] - CK-12 Biology for High School- 2.28 Electron Transport, Figure 2
| |
|
| |
| :::::: [[File:Hyperphysics CORRECTION.png|400px]]
| |
| :::: '''Website 24''': [http://hyperphysics.phy-astr.gsu.edu/hbase/Biology/Complex1.html hyperphysics]
| |
|
| |
| :::::: [[File:Labster Theory CORRECTION.png|400px]]
| |
| :::: '''Website 25''': [https://theory.labster.com/Electron_Transport_Chain/ Labster Theory]
| |
|
| |
| :::::: [[File:Nau.edu CORRECTION.png|400px]]
| |
| :::: '''Website 26''': [https://www2.nau.edu/~fpm/bio205/u4fg36.html nau.edu]
| |
|
| |
| :::::: [[File:Quizlet CORRECTION.png|400px]]
| |
| :::: '''Website 27''': [https://quizlet.com/245664214/electron-transport-chain-facts-of-cell-respiration-diagram/ Quizlet]
| |
|
| |
| :::::: [[File:ScienceDirect CORRECTION.png|400px]]
| |
| :::: '''Website 28''': [https://www.google.com/imgres?imgurl=https%3A%2F%2Fars.els-cdn.com%2Fcontent%2Fimage%2F3-s2.0-B9780128008836000215-f21-07-9780128008836.jpg&imgrefurl=https%3A%2F%2Fwww.sciencedirect.com%2Ftopics%2Fengineering%2Felectron-transport-chain&tbnid=g3dD4u8Tvd6TWM&vet=12ahUKEwjc9deUprT9AhVxhv0HHXZbAd0QMygCegUIARDBAQ..i&docid=Moj_2_W0OpUDcM&w=632&h=439&q=FADH2%20is%20the%20substrates%20of%20Complex%20II&client=firefox-b-d&ved=2ahUKEwjc9deUprT9AhVxhv0HHXZbAd0QMygCegUIARDBAQ ScienceDirect]
| |
|
| |
| :::::: [[File:ScienceFacts CORRECTION.png|400px]]
| |
| :::: '''Website 29''': [https://www.sciencefacts.net/electron-transport-chain.html ScienceFacts]
| |
|
| |
| :::::: [[File:SNC1D CORRECTION.png|400px]]
| |
| :::: '''Website 30''': [https://sbi4uraft2014.weebly.com/electron-transport-chain.html SNC1D - BIOLOGY LESSON PLAN BLOG]
| |
|
| |
| :::::: [[File:Unm.edu CORRECTION.png|400px]]
| |
| :::: '''Website 31''': [https://www.unm.edu/~lkravitz/Exercise%20Phys/ETCstory.html unm.edu]
| |
|
| |
| :::::: [[File:Wikimedia ETC CORRECTION.png|400px]]
| |
| :::: '''Website 9''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - By User:Rozzychan CC BY-SA 2.5, via Wikimedia Commons
| |
| :::: '''Website 32''': [https://commons.wikimedia.org/wiki/File:Mitochondrial_electron_transport_chain.png Wikimedia]
| |
|
| |
| :::::: [[File:YouTube Dirty Medicine Biochemistry CORRECTION.png|400px]]
| |
| :::: '''Website 33''': [https://www.google.com/imgres?imgurl=https%3A%2F%2Fi.ytimg.com%2Fvi%2FLsRQ5_EmxJA%2Fmaxresdefault.jpg&tbnid=6w-0DVPMw7vOdM&vet=12ahUKEwjw2YO5--T9AhUwpCcCHduuDVgQMygDegUIARDzAQ..i&imgrefurl=https%3A%2F%2Fwww.youtube.com%2Fwatch%3Fv%3DLsRQ5_EmxJA&docid=bZxQYNch1Ys-VM&w=1280&h=720&q=electron%20transport%20chain&hl=en-US&client=firefox-b-d&ved=2ahUKEwjw2YO5--T9AhUwpCcCHduuDVgQMygDegUIARDzAQ YouTube Dirty Medicine Biochemistry] - Uploaded 2019-07-18
| |
|
| |
| :::::: [[File:YouTube sciencemusicvideos CORRECTION.png|400px]]
| |
| :::: '''Website 34''': [https://www.google.com/imgres?imgurl=https%3A%2F%2Fi.ytimg.com%2Fvi%2FVER6xW_r1vc%2Fmaxresdefault.jpg&tbnid=Brshl0oN9LyYnM&vet=12ahUKEwjjlKSKpOX9AhWjmycCHbvGC34QMygWegUIARDWAQ..i&imgrefurl=https%3A%2F%2Fwww.youtube.com%2Fwatch%3Fv%3DVER6xW_r1vc&docid=VgTgrLf24Lzg4M&w=1280&h=720&itg=1&q=FADH2%20is%20the%20substrates%20of%20Complex%20II&hl=en&client=firefox-b-d&ved=2ahUKEwjjlKSKpOX9AhWjmycCHbvGC34QMygWegUIARDWAQ YouTube sciencemusicvideos] - Uploaded 2014-08-19
| |
|
| |
| :::::: [[File:ThoughtCo-Getty Images CORRECTION.png|400px]]
| |
| :::: '''Website 15''': [https://www.thoughtco.com/electron-transport-chain-and-energy-production-4136143 ThoughtCo] - extender01 / iStock / Getty Images Plus
| |
| :::: '''Website 17''': [https://www.dreamstime.com/royalty-free-stock-photography-electron-transport-chain-illustration-oxidative-phosphorylation-image36048617 dreamstime]
| |
|
| |
|
| |
| == CII and fatty acid oxidation ==
| |
|
| |
| :::::::: [[File:SUIT-catg F.jpg|300px|F-junction|link=Fatty acid oxidation pathway control state]] [[File:Wang 2019 Fig8.png|300px|link=Wang Y 2019 J Biol Chem]]
| |
| :::: Fatty acid oxidation requires electron transferring flavoprotein CETF and CI for electron entry into the Q-junction ([[Gnaiger_2020_BEC_MitoPathways |Gnaiger 2020]]; [[Wang Y 2019 J Biol Chem |Wang et al 2019]]; see figures on the right).
| |
|
| |
| :::::: [[File:Missaglia 2021 Crit Rev Biochem Mol Biol CORRECTION.png|600px]]
| |
| :::: When FADH<sub>2</sub> is erroneously shown as a substrate of CII <span style="color:#FF0000">(1)</span>, a role of CII in fatty acid oxidation is suggested as a consequence <span style="color:#FF0000">(2)</span>.
| |
|
| |
| :::: '''Website 35''': [https://conductscience.com/electron-transport-chain/ Conduct Science]: "In Complex II, the enzyme succinate dehydrogenase in the inner mitochondrial membrane reduce FADH<sub>2</sub> to FAD<sup>+</sup>. Simultaneously, succinate, an intermediate in the Krebs cycle, is oxidized to fumarate." - Comments: FAD does not have a postive charge. FADH<sub>2</sub> is the reduced form, it is not reduced. ''And again:'' In CII, FAD is reduced to FADH<sub>2</sub>.
| |
|
| |
| :::::: [[File:Expii-Gabi Slizewska CORRECTION.png|600px]]
| |
| :::: '''Website 9''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Image source: By Gabi Slizewska. The ambiguity in the graphical representation is solidified as an error in the text: ''In the second step, Complex II receives electrons from FADH<sub>2</sub>, oxidizing it to FAD.''
| |
|
| |
| ::::* "Since mitochondrial Complex II also participates in the oxidation of fatty acids (6), .." (quote from [[Lemmi 1990 Biochem Med Metab Biol |Lemmi et al 1990]]).
| |
| ::::::* ''Ref 6'': Tzagoloff A (1982) Mitochondria. Plenum, New York.
| |
| ::::::* This quote is ambiguous. The textbook by [[Tzagoloff 1982 Plenum Press |Tzagoloff (1982)]] represents fatty acid oxidation in figures and text without involvement of CII. Oxidation of acetyl-CoA, however, proceeds in the TCA cycle into which CII is integrated.
| |
|
| |
| :::::: [[File:FAO-CII Medical Biochemistry Page.jpg|400px|right|link=https://themedicalbiochemistrypage.org/oxidative-phosphorylation-related-mitochondrial-functions/]]
| |
| :::: '''Website 36''': [https://themedicalbiochemistrypage.org/oxidative-phosphorylation-related-mitochondrial-functions/ The Medical Biochemistry Page] - Figure and text go together: "''In addition to transferring electrons from the FADH<sub>2</sub> generated by SDH, complex II also accepts electrons from the FADH<sub>2</sub> generated during fatty acid oxidation via the fatty acyl-CoA dehydrogenases and from mitochondrial glycerol-3-phosphate dehydrogenase (GPD2) of the glycerol phosphate shuttle''".
| |
|
| |
| :::: '''Website 37''': [https://www.chem.purdue.edu/courses/chm333/Spring%202013/Lectures/Spring%202013%20Lecture%2037%20-%2038.pdf CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna] - Acyl-CoA dehydrogenase is listed under 'Electron transfer in Complex II'.
| |
|
| |
|
| |
| {{MitoPedia concepts | | {{MitoPedia concepts |
| |mitopedia concept=MiP concept | | |mitopedia concept=MiP concept |