Yepez 2018 PLOS One
| Yépez VA, Kremer LS, Iuso A, Gusic M, Kopajtich R, Koňaříková E, Nadel A, Wachutka L, Prokisch H, Gagneur J (2018) OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. https://doi.org/10.1371/journal.pone.0199938 |
» PLOS ONE 13:e0199938. PMID: 29995917 Open Access
Yepez VA, Kremer LS, Iuso A, Gusic M, Kopajtich R, Konarikova E, Nadel A, Wachutka L, Prokisch H, Gagneur J (2018) PLOS ONE
Abstract: The accurate quantification of cellular and mitochondrial bioenergetic activity is of great interest in medicine and biology. Mitochondrial stress tests performed with Seahorse Bioscience XF Analyzers allow the estimation of different bioenergetic measures by monitoring the oxygen consumption rates (OCR) of living cells in multi-well plates. However, studies of the statistical best practices for determining aggregated OCR measurements and comparisons have been lacking. Therefore, to understand how OCR behaves across different biological samples, wells, and plates, we performed mitochondrial stress tests in 126 96-well plates involving 203 fibroblast cell lines. We show that the noise of OCR is multiplicative, that outlier data points can concern individual measurements or all measurements of a well, and that the inter-plate variation is greater than the intra-plate variation. Based on these insights, we developed a novel statistical method, OCR-Stats, that: i) robustly estimates OCR levels modeling multiplicative noise and automatically identifying outlier data points and outlier wells; and ii) performs statistical testing between samples, taking into account the different magnitudes of the between- and within-plate variations. This led to a significant reduction of the coefficient of variation across plates of basal respiration by 45 % and of maximal respiration by 29 %. Moreover, using positive and negative controls, we show that our statistical test outperforms the existing methods, which suffer from an excess of either false positives (within-plate methods), or false negatives (between-plate methods). Altogether, this study provides statistical good practices to support experimentalists in designing, analyzing, testing, and reporting the results of mitochondrial stress tests using this high throughput platform.
Selected quotes
- Power analysis
- By doing power analysis, we showed that our method is able to detect relative differences of 10 % - 15 %, and that the minimum number of well replicates per biological sample in a 96-well plate should be 12.
- Furthermore, our study provides experimental design guidance by i) showing that between-plate variation largely dominates within-plate variation, implying that it is important to seed the same cell lines in multiple plates, and ii) providing estimates of variances within and between plates for each bioenergetic measure allowing for statistical power computations.
- Using data from the controls NHDF, we found that the variability between plates in all the intervals is much larger than that between wells (S2 Table and S5 Fig). .. These observations show the importance of basing conclusions from observations across multiple plates and for seeding a control cell line on every plate.
- Assuming three plates per comparison and 16 wells per plate, these standard deviations allow detecting relative differences of 10 % to 15 % depending on the considered log OCR ratios differences for significance level of 5 % (Fig 6, right y-axis, Materials and methods). Relative differences of 10 % to 15 % are in line with reported detected variations in the literature which we found to be as low as 12%-30% [30–32]. This analysis also suggests to seed at least 12 wells per biological sample per plate, since we observed increased standard deviations of the residuals for numbers of wells smaller than 12.
- One approach to evaluate samples measured in multiple plates is to perform a Wilcoxon test on the ED values averaged per plate (across-plate ED, Materials and methods). However, this requires at least five plate replicates in order to obtain significant results.
- Comment: Evaluation of variability between plates requires parallel measurements in different instruments, to eliminate differences due to measurements on different days.
- Systematic effects - bias
- We investigated whether there were positional effects and found that OCR measurements are lower in the edges by a median of up to 13.1% (S4A and S4B Fig). However, these positional effects were consistent across intervals (S4A and S4B Fig). .. . Practitioners could avoid all four edges and not only the four corners as typically done.
- .. we observed systematic effects in OCR between wells (e.g., OCR values of well C6 are among the highest, while OCR values of well B5 are among the lowest at all the time points; Fig 2A).
- Comment: All four edges contain 36 wells, leaving 60 available wells on a 96 well plate. Consider 12 wells for controls, then 48 wells remain for the experimental groups. With 12 repeats per group, four experimental groups can be tested per 96 well-plate, not considering outliers.
- Normalization
- Strikingly, dividing OCR by cell count led to a higher coefficient of variation (standard deviation divided by the mean) between the replicate wells than without that correction (S1B Fig).
- Since there is a remaining systematic effect across intervals at the plate level (Fig 3C) and because of the plate-interval effects, we recommend using ratios of OCR levels (i.e. differences in the logarithmic scale) (Table 2).
- Outliers
- Around 16.5 % of all valid wells detected as outliers.
- The wells for which the median OCR level did not follow the expected order, namely, median [OCR(Int3)] > median[OCR(Int1)] > median[OCR(Int2)] > median[OCR(Int4)] (Fig 1A), were discarded (977 wells, 10.47 %). .. In addition, we excluded contaminated wells and wells in which the cells got detached (461 wells, 4.94 %) from the analysis.
- Selection of time points in each respiratory state
- Extreme differences (default) method to compute bioenergetic measures. - On every plate independently, for each well, in interval 1 take the OCR corresponding to the last measurement, in intervals 2 and 4 take the minimum, and in interval 3 the maximum OCR value [19]. Then, use the corresponding differences to estimate the bioenergetic measures. Report the results per patient as the mean across wells plus standard deviation or standard error, separately for each plate.
Respiratory states
- Interval 1: ROUTINE - take measurement 3
- Interval 2: LEAK (Omy) - take the minimum
- Interval 3: ET (Uncoupler) - take the maximum
- Interval 4: Rbl (Rot&Ama) - take the minimum
Cited by
- Gnaiger E (2021) Bioenergetic cluster analysis – mitochondrial respiratory control in human fibroblasts. MitoFit Preprints 2021.8. https://doi.org/10.26124/mitofit:2021-0008
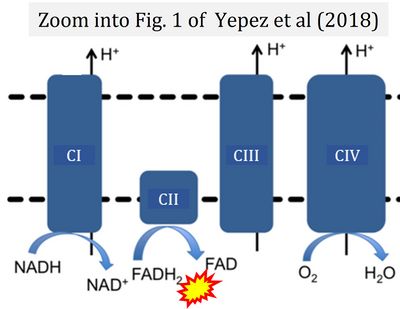
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Labels: MiParea: Instruments;methods
Preparation: Intact cells
Coupling state: LEAK, ROUTINE, ET
MitoFit 2021 BCA, PLoSONE2022ace-sce, MitoFit2022QC