MiPNet08.09 CellRespiration
| High-resolution respirometry and coupling-control protocol with living cells: ROUTINE, LEAK, ET-pathway, ROX. |
» ![]() »Versions
»Versions
Oroboros (2016-08-11) Mitochondr Physiol Network
Abstract: Doerrier C, Gnaiger E (2003-2016) High-resolution respirometry and coupling-control protocol with living cells: ROUTINE, LEAK, ET-pathway, ROX. Mitochondr Physiol Network 08.09(11):1-8.
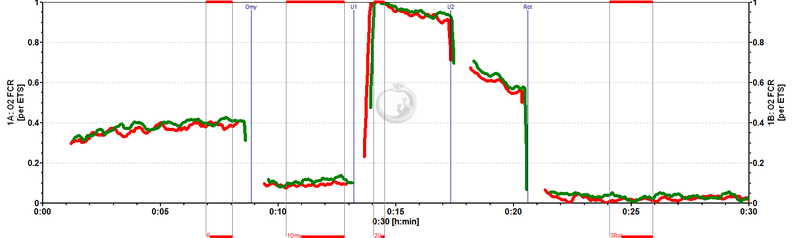
An experiment on respiration of living cells is reported from an O2k-Workshop on high-resolution respirometry. Leukemia cells were incubated at a density of 1 million cells/ml in 2 ml culture medium in two O2k-Chambers operated in parallel. Cellular ROUTINE respiration, JR, resulted in volume-specific oxygen consumption of 20 pmol·s-1·ml-1. Oxygen concentration changed by merely 6.4 and 6.5 µM in the two O2k-Chambers over a period of 5 min (<1% air saturation per minute). Inhibition by oligomycin (JL), and rotenone (residual oxygen consumption, JROX; after uncoupling) reduced respiration to 5 and 1 pmol·s-1·ml-1, while inducing the noncoupled state by the uncoupler FCCP revealed the capacity of the Electron transfer-pathway (ET-pathway) at JE of 50 pmol·s-1·ml-1. The ROUTINE control ratio, R/E, was 0.4 (uncoupling control ratio, UCR=E/R=2.5), and the LEAK control ratio, L/E, was 0.1 (E/L=12.0). This indicates tight coupling of OXPHOS, and a large ET-pathway excess capacity over ROUTINE respiration. The net ROUTINE control ratio, netR=(R-L)/E was 0.30, indicating that 30% of ET-pathway capacity was activated for ATP production.
Automatic correction for instrumental background amounted to 13% for ROUTINE respiration, but to >50% and 180% for JL and JROX, respectively, illustrating the importance of real-time correction. The experiment illustrates the sensitivity and resproducibility of high-resolution respirometry with the OROBOROS O2k. Calibrations and routine corrections provide the basis of the high accuracy required for mitochondrial respiratory physiology. Real-time analyses were performed, combining high-resolution with instant diagnostic information. In this update graphs are presented illustrating some features of DatLab.
- » Product: Oroboros O2k, O2k-Catalogue
• Bioblast editor: Gnaiger E • O2k-Network Lab: AT_Innsbruck_Oroboros
Labels: MiParea: Respiration, Instruments;methods
Organism: Human
Tissue;cell: Blood cells, Lymphocyte
Preparation: Intact cells
Coupling state: LEAK, ROUTINE, ET
HRR: Oxygraph-2k, O2k-Protocol
O2k-Demo, O2k-Core, SUIT-003 O2 ce D009, SUIT-003, SUIT-006 General O2 pce D053, Leukemia
» MitoPedia: Respiratory states
![]()
![]()
![]()

Flux control ratios (FCR) normalized to state ET-pathway (E) in chambers A and B (superimposed). ROUTINE respiration in these cells is regulated at 0.4 of ET capacity (R/E). Oligomycin (Omy) inhibits respiration to 0.1 ET capacity (L/E).
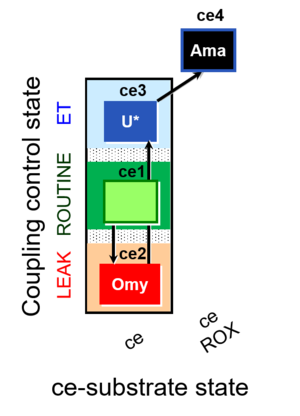
Coupling-control protocol
- ceCCP: RLE 1ce;2ceOmy;3ceU-
- » Instructions for using templates for data evaluation are given in MiPNet08.09 (see above) and MiPNet10.04 CellRespiration.
ceCCP states
| Respiratory state | Mark names | Explanations |
|---|---|---|
| R | 1ce | ROUTINE |
| L | 2ceOmy | Oligomycin |
| E | 3ceU | Uncoupler titration |
| ROX | 4ceRotAma | Rotenone, Antimycin A, residual oxygen consumption |
DatLab-Analysis templates and DatLab Demo Files
DatLab 7
- Calibration-File: MiPNet08.09_CellRespiration_Calib.DLD
- Demo-File: MiPNet08.09_CellRespiration.DLD
- DatLab-Analysis template: CCP02.xlsx
DatLab 6
- Calibration-File: MIPNET08.09_2003-03-29_P1-01_CALIB.DLD
- Demo-File: MIPNET08.09_2003-03-29_P1-02_CELLS.DLD
- Background Excel demo and template: MiPNet08.09_O2k-Background_Cells.xlsx
- DatLab-Analysis template: MiPNet08.09_O2k-Analysis_Cells.xlsx
DatLab 5
- Calibration-File: MIPNET08.09_2003-03-29_P1-01_CALIB.DLD
- Demo-File: MIPNET08.09_2003-03-29_P1-02_CELLS.DLD
- Background Excel demo and template: O2k-Background_Cells_0809.xlsx
- DatLab-Analysis template: O2k-Analysis_Cells_0809.xlsx