Ma 2020 Sci Rep
| Ma Y, Wang W, Devarakonda T, Zhou H, Wang XY, Salloum FN, Spiegel S, Fang X (2020) Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci Rep 10:1450. |
Ma Y, Wang W, Devarakonda T, Zhou H, Wang XY, Salloum FN, Spiegel S, Fang X (2020) Sci Rep
Abstract: Fatty acid oxidation (FAO) is a key bioenergetic pathway often dysregulated in diseases. The current knowledge on FAO regulators in mammalian cells is limited and sometimes controversial. Previous FAO analyses involve nonphysiological culture conditions or lack adequate quantification. We herein described a convenient and quantitative assay to monitor dynamic FAO activities of mammalian cells in physiologically relevant settings. The method enabled us to assess various molecular and pharmacological modulators of the FAO pathway in established cell lines, primary cells and mice. Surprisingly, many previously proposed FAO inhibitors such as ranolazine and trimetazidine lacked FAO-interfering activity. In comparison, etomoxir at low micromolar concentrations was sufficient to saturate its target proteins and to block cellular FAO function. Oxfenicine, on the other hand, acted as a partial inhibitor of FAO. As another class of FAO inhibitors that transcriptionally repress FAO genes, antagonists of peroxisome proliferator-activated receptors (PPARs), particularly that of PPARα, significantly decreased cellular FAO activity. Our assay also had sufficient sensitivity to monitor upregulation of FAO in response to environmental glucose depletion and other energy-demanding cues. Altogether this study provided a reliable FAO assay and a clear picture of biological properties of potential FAO modulators in the mammalian system.
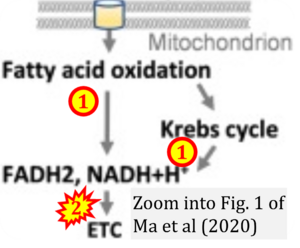
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Cited by
- Silva et al (2021) Off-target effect of etomoxir on mitochondrial Complex I. MitoFit Preprints 2021. (in preparation)
Labels:
Enzyme: Complex II;succinate dehydrogenase
Pathway: F
MitoFit 2021 Etomoxir